-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Sequences Analysed
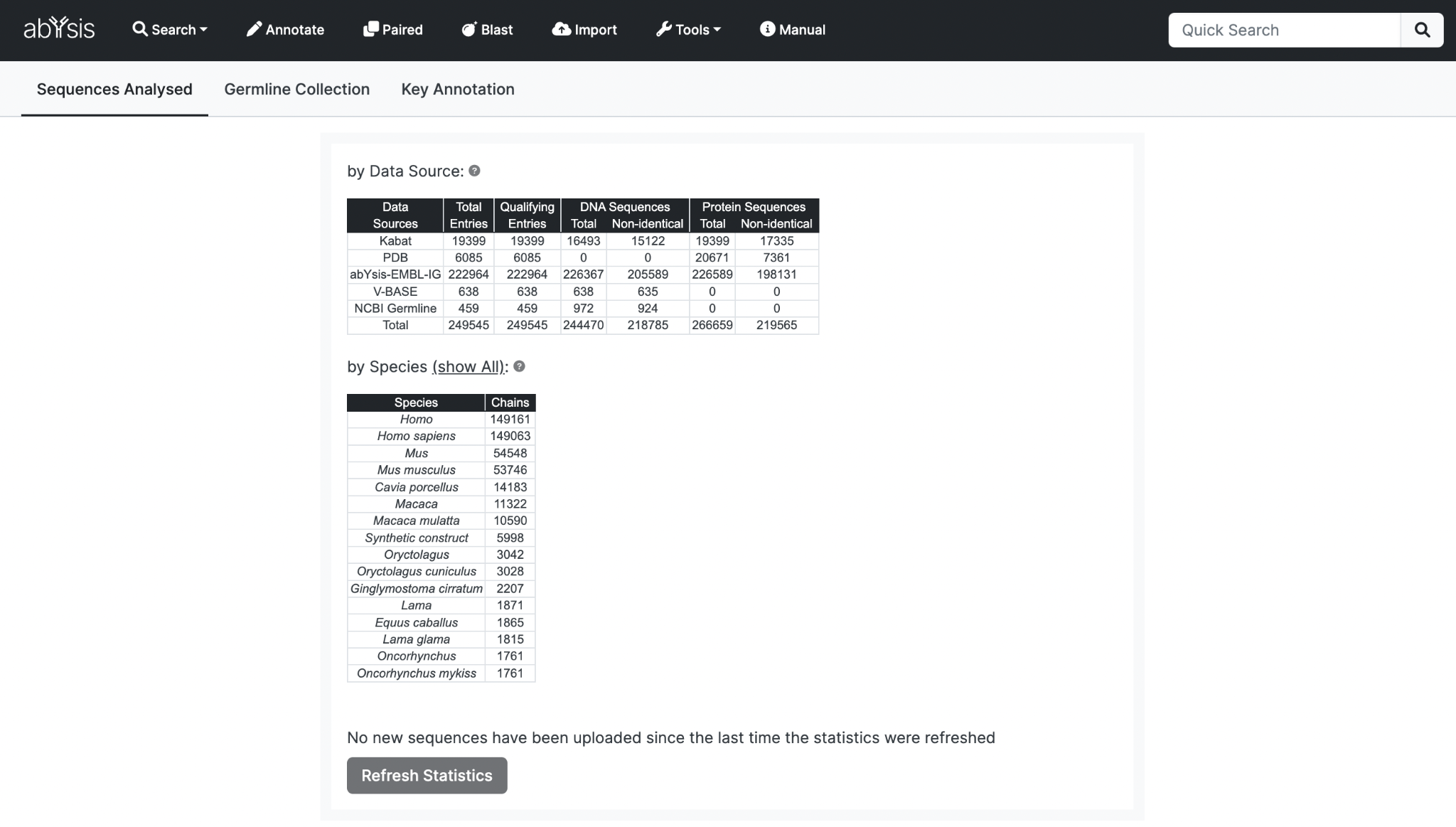
Data Source Summary
Data Sources Entries from public resources or Import function.
Total Entries Total number of all entries for each Data Source - even if non-antibody.
Qualifying Entries Those assessed as containing immunoglobulin sequence data based on textual annotations such as keywords and descriptions.
More than one protein sequence may be available in a Qualifying Entry so total number of proteins sequences may be greater than the number of qualifying entries.
- EMBL; single DNA entry may have multiple protein
- PDB; may be more than one protein chain.
Also abYsis splits antibodies such as scFVs into separate entries. So for EMBL entries, DNA and protein sequences are split. For PDB, scFV protein sequence chains (and coordinates may be split).
Note: abYsis-EMBL-IG data source is compiled automatically based on sequence similarity to known, curated immunoglobulin sequences.
- Several thousand representative immunoglobulin query sequences of known structure have been used to identify immunoglobulins in the EMBL database whilst also minimising mis-selection of T-cell receptors.
Most Frequent Species
Top 15 Shown by default.
Note: Number of stored chains may be higher than the number retrievable in searches as not all sequences will be successfully numbered.
Chain: Basic data entity in abYsis. A chain may be a DNA or protein sequence, or both.
