-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Sequence Import
The commercial version of abYsis allows users to import sequences into the abYsis database and process them for analysis.
Select Import on the abYsis main menu:

On first usage you will most likely be presented with this screen where the address will the where your IT team have installed your local version of abYsis.
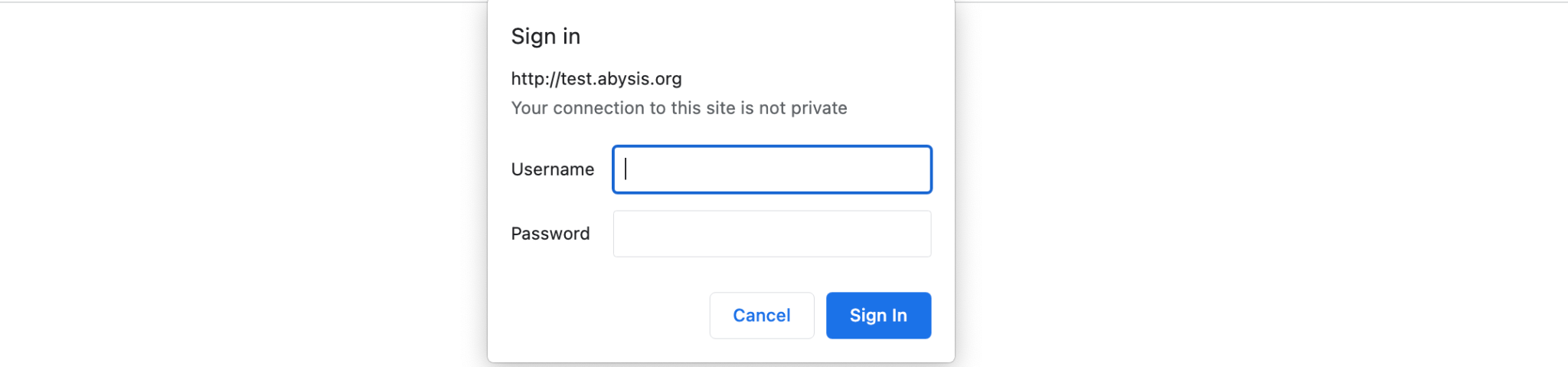
They will have determined what the Username & Password should be during the installation process. This allows companies to restrict who can import sequences into abYsis whilst still providing broader access to other users.
Note: You follow the abYsis-specific Fasta format described in this document for trouble-free access.
Once you have successfully entered you will see the following screen;
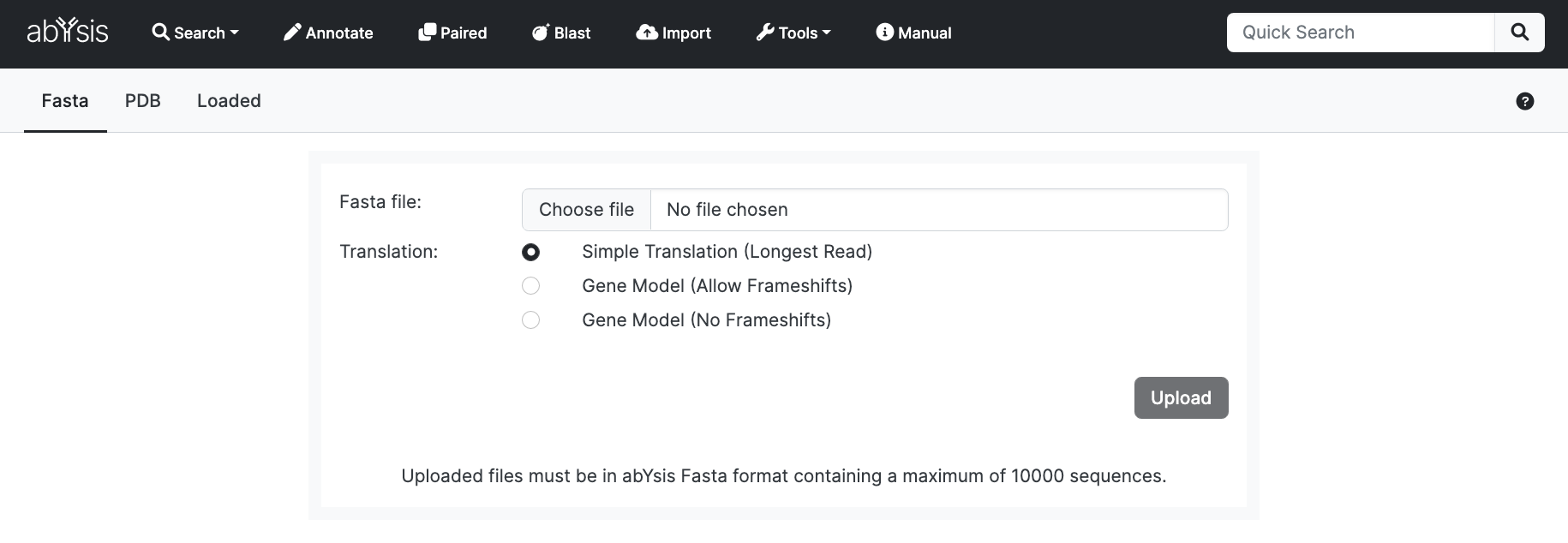
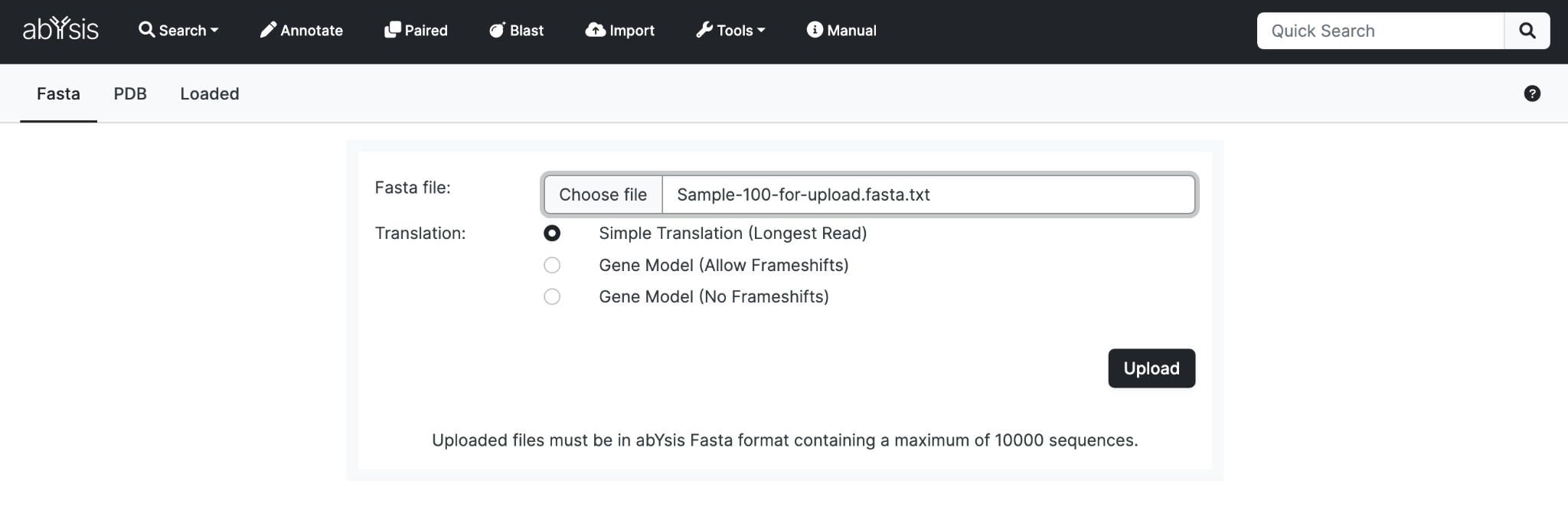
Select File
From the Upload tab, select a file from your local computer. It must be in the Fasta based format described later
Translation method:
Unless you have particular reason to select another, we suggest simple translation. See Annotate section for more detail.
Upload
Click Upload to upload your dataset. You can monitor progress on the Loaded tab.
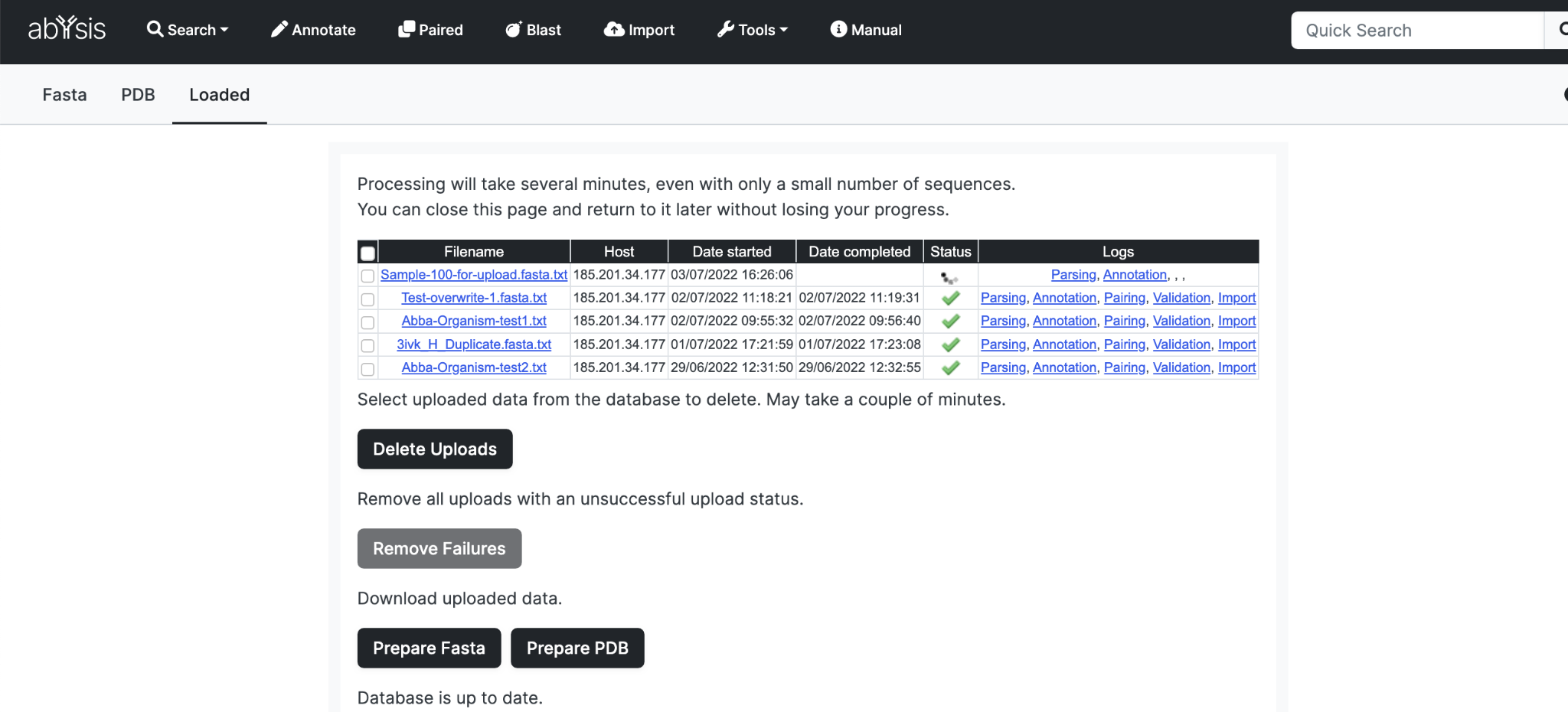
An active job is indicated by a spinning icon. (Currently, the system only permits one active job.)
Successful processing is indicated by a green tick.
A red cross indicates and issue and you should look at the log files for feedback and issues with your upload.
Once your data is in the database the web pages will reflect the updated information.
Options
Note: Do not run more than one of the following options at the same time. Wait for each one to finish before progressing. In the event that more than one option is accidentally activated you should wait the allotted time and then manually refresh the page as it is possible only the first completed task will automatically update the page.
Delete Uploads
You can remove uploads from the database using the Delete Uploads button. Select which entries to delete and click the button. It is recommended not to do this action whilst a statistics and frequencies rebuild is occuring.
Remove failures
If any data fails to upload then a failed entry may exist in the table. To remove all unsuccessful uploads click the Remove failures button.
Download Uploaded Data
It is possible to download all the uploaded sequences. This is a good way to back-up your uploaded sequences and transfer them to a new installation.
Click the Prepare Fasta and wait.
When ready the button Download Fasta will appear.
Rebuild Statistics and Frequencies
This will update the Statistics tab and reflect uploads that have been made since the last Rebuild. This will also update the amino acid frequencies throughout the site. This action can take up to an hour on older servers. Please be patient.
Note: You may need to force refresh/reload of pages if you browser has cached an older version.
Optimise Database
After uploading more than 10,000 sequences it can be beneficial to optisise the database. The Optimise button will only be available when 10,000 new sequences have been uploaded. Database will be slower for a couple of hours while optimisation occurs so you are recommended to do overnight or when system usage is lower.
abYsis Fasta Format
Input file must conform to a specific format.
The Title line must start with a '>' character, followed by six fields separated by vertical bars '|' as follows:
>accession|partner|species|datasource|chaintype|moleculetype
- accession An accession code for the chain (required)
- partner An accession code for the partner chain (required only for pairing)
- species A Latin species name, lower case (optional but recommended. You can make your own species if you wish to keep data separate)
- datasource A data source name (required)
- chaintype The chain type: 'heavy' or 'light' (optional for unpaired, required for paired)
- moleculetype The sequence type 'dna' or 'protein' (required)
An example:
>001485|007059|homo sapiens|mydata|heavy|protein QVQLKESGPGLVPSSLSCTVSFSTSGVWVRPPGKGLEWLMIWSGGSTVNAAFISRLSISK DNSKSVFMNSLQANDTAIYYCARDYGYQGTLTVSA
Further notes:
- While each file may only contain <% $maxUploadSeqs %> sequences, you may upload multiple files.
- Accessions must be unique for a given data source. Paired chains must have the same data source and be in the same file.
- You may have a mix of paired and unpaired chains in the same file.
- Chain types must be provided for paired chains.
- Pairing information must be consistent within the file. If the new data replaces or updates data_source/accessions that are already loaded in the database, pairing information for the chains involved will be deleted from the database prior to loading the new data.
All fields must be present but optional fields can be left empty.
For example, if a chain is unpaired, leave the partner accession field empty, like this:
>001485||homo sapiens|mydata|heavy|protein QVQLKESGPGLVPSSLSCTVSFSTSGVWVRPPGKGLEWLMIWSGGSTVNAAFISRLSISK DNSKSVFMNSLQANDTAIYYCARDYGYQGTLTVSA
You may provide both DNA and/or protein sequence data for a given accession.
If providing both, repeat the title line (exactly) except for the final field (protein or dna).
>001485|007059|homo sapiens|mydata|heavy|protein QVQLKESGPGLVPSSLSCTVSFSTSGVWVRPPGKGLEWLMIWSGGSTVNAAFISRLSISK DNSKSVFMNSLQANDTAIYYCARDYGYQGTLTVSA >001485|007059|homo sapiens|mydata|heavy|dna caggtgcagctgaaggagtcaggacctggcctagtgccctcagcctgtcctgcacagtct ctttctcactagcggtgttgggttcgccctccaggaaagggtctggagtggctgatgatatg gagtggtggaagcacagtcaatgcagctttcatatccagactgagcatcagcaaggaca attccaagagcgtttttatgaacagtctgcaagctaatgacacagccatatattactgtgcca gagactatgggtaccaagggactctgactgtctctgca
Other points:
- Accessions must be unique for a given data source
- Paired chains must have the same data source
- You may have a mix of paired and unpaired chains in the same file
- The chain types must be provided for paired chains
- Pairing information must be consistent within the file
Avoiding Errors
Only use pairing information if you really require it
Example 1.
This file only has one entry, but it will load correctly because there is no accession in the second field that describes heavy chain/light chain pairing. You do not require both protein and DNA, the system will work with just one of these.
>140665||mus musculus|mydata|heavy|protein QVQLKESGPGLVPSSLSCTVSFSTSGVWVRPPGKGLEWLMIWSGGSTVNAAFISRLSISK DNSKSVFMNSLQANDTAIYYCARDYGYQGTLTVSA >140665||mus musculus|mydata|heavy|dna caggtgcagctgcaggagtctggacctggcctagtgcagccctcacagagcctgtccatcacct gcacagtctctggtttctcattaactagctatggtgtacactgggttcgccagtctccaggaaaggg tctggagtggctgggagtgatatggagtggtggaagcacagactataatgcagctttcatatcca gactgagcatcagcaaggacaattccaagagccaagttttctttaaaatgaacagtctgcaagct aatgacacagccatatattactgtgccagaaacgcttactggggccaagggactctggtcactgt ctctgca
Example 2.
This file also has only one entry but loading will fail because there is an accession for pairing file (020167) yet no sequence with accession 020167 in the upload file.
>140666|020167|mus musculus|johnfile|heavy|protein QVQIKESGPGLVPSSLSCTVSFSTSGVWVRPPGKGLEWLMIWSGGSTVNAAFISRLSISK DNSKSVFMNSLQANDTAIYYCARDYGYQGTLTVSA >140666|020167|mus musculus|johnfile|heavy|dna aggtgcagctgcaggagtctggacctggcctagtgcagccctcacagagcctgtccatcacct gcacagtctctggtttctcattaactagctatggtgtacactgggttcgccagtctccaggaaaggg tctggagtggctgggagtgatatggagtggtggaagcacagactataatgcagctttcatatcca gactgagcatcagcaaggacaattccaagagccaagttttctttaaaatgaacagtctgcaagct aatgacacagccatatattactgtgccagaaacgcttactggggccaagggactctggtcactgt ctctgca
Example 3.
This contains two sequences that will load correctly because the pairing information refers to another sequence in the upload file.
>Z-AAA50198|Z-AAA50199|mus musculus|nishikawadata|heavy|protein LESGAELVKPGASVKLSCKASGYTFSSYWMHWVKQRPGQGLEWIGEIHPSNGLTNYNEKF KSKATLTVDKSSSTAYMQLSSLTSEDSAVYYCAKGKELGRFAYWGQGTLVTVSA >Z-AAA50198|Z-AAA50199|mus musculus|nishikawadata|heavy|dna ctcgagtctggggctgaactggtgaagcctggggcttcagtgaagttgtcctgcaaggcttct ggctacaccttcagtagttactggatgcactgggtgaagcagaggcctggacaaggacttg agtggattggagagattcatcctagcaacggtcttactaactataatgagaagttcaagagt aaggccacattgactgtagacaaatcctccagcacagcctacatgcaactcagcagcctg acatctgaggactctgcggtctattactgtgcaaaagggaaggaactgggacggtttgcgt actggggccaagggactctggtcactgtctctgca >Z-AAA50199|Z-AAA50198|mus musculus|nishikawadata|light|protein TQSPASLAVSLGQRATISCRASESVDSYGNSFMHWYQQKPGQPPKLLIYLASNLESGVPA RFSGSGSRTDFTLTIDPVEADDAATYYCQQNNEDPYTFGGGTKLEIKRA >Z-AAA50199|Z-AAA50198|mus musculus|nishikawadata|light|dna acccagtctccagcttctttggctgtgtctctagggcagcgggccaccatatcctgcagagcc agtgaaagtgttgatagttatggcaatagttttatgcactggtaccagcagaaaccaggaca gccacccaaactcctcatctatcttgcatccaacctagaatctggggtccctgccaggttcagt ggcagtgggtctaggacagacttcaccctcaccattgatcctgtggaggctgatgatgctgca acctattactgtcagcaaaataatgaggatccgtacacgttcggaggggggaccaagctgg aaataaaacgggct
Example 4.
Something like this will also load as there is still sufficient pairing information in the upload file.
>140667|020169|mus musculus|andrew-ig|heavy|protein QVQIKESGPGLVPSSLSCTVSFSTSGVWVRPPGKGLEWLMIWSGGSTVNAAFISRLSISK DNSKSVFMNSLQANDTAIYYCARDYGYQGTLTVSA >140667|020169|mus musculus|andrew-ig|heavy|dna aggtgcagctgcaggagtctggacctggcctagtgcagccctcacagagcctgtccatcacct gcacagtctctggtttctcattaactagctatggtgtacactgggttcgccagtctccaggaaaggg tctggagtggctgggagtgatatggagtggtggaagcacagactataatgcagctttcatatcca gactgagcatcagcaaggacaattccaagagccaagttttctttaaaatgaacagtctgcaagct aatgacacagccatatattactgtgccagaaacgcttactggggccaagggactctggtcactgt ctctgca >020169|140667|mus musculus|andrew-ig|light|dna gtgcagatcaaggagagcggccccggcctggtgcccagcagcctgagctgcaccgtgagc ttcagcaccagcggcgtgtggatcaggccccccggcaagggcctggagtggctgatgatct ggagcggcggcagcaccgtgaacgccgccttcatcaccaagctgagcatcagcagggac aacagcaagagcgtgttcatgcagagcctgcaggccaacgacaccgccatctactactgcg ccagggagtacggctaccagggcaccctgaccgtgagcgc