-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Sequence
 Use sequence fragments, motifs and residues as well as specific positions to search for sequences in the database
Use sequence fragments, motifs and residues as well as specific positions to search for sequences in the database
Note:
- Terms may be specified separately for heavy and light chains.
- If you specify terms for both heavy and light chains in this section or elsewhere in the form, the search will be restricted to paired sequences.
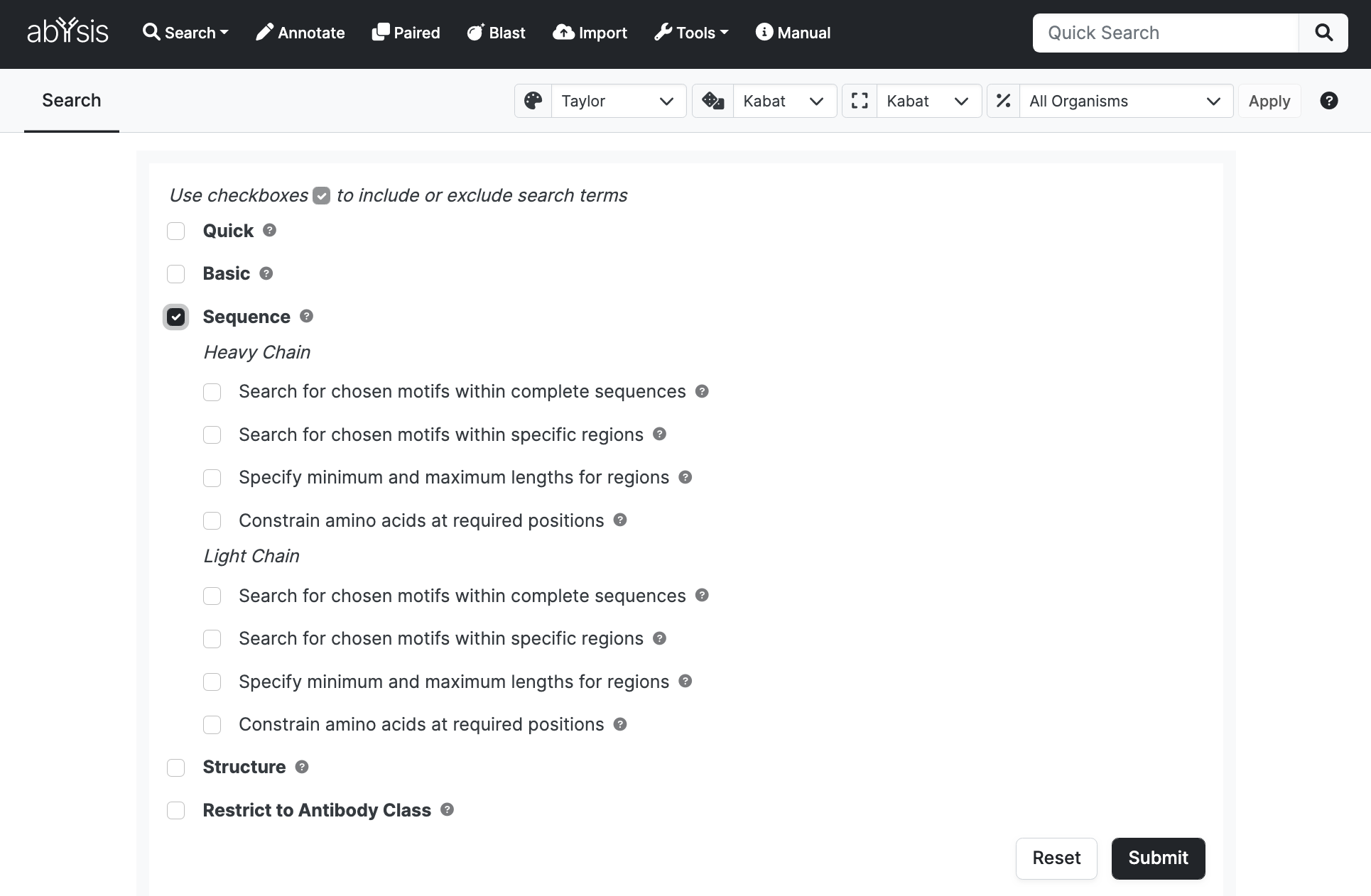
Search for chosen motifs within complete sequences
 Note: This option usually runs on the full sequence, not just the numbered region. Therefore some hits might not be displayed in the numbered alignment.
Note: This option usually runs on the full sequence, not just the numbered region. Therefore some hits might not be displayed in the numbered alignment.
To restrict to numbered region see ‘Search for chosen motifs within specific regions’
Restrict the search to sequences which contain particular fragments or motifs.
- Select contains in the dropdown to specify an exact sequence fragment which must be present and enter the sequence (e.g. EDGV) in the text box.
- Select ~ (regex) in the dropdown and a enter a regular expression e.g. EDG(V|L) to indicate glu-asp-gly followed by val or leu or AD[^VL] to indicate ala-asp followed by residues other than val or leu. For other ideas of how Regex work for sequences, look at the Post Translational Modificiations (PTMs) section which are all defined by Regex. Note that for Regex you must select Regex from the dropdown menu.
The search is case insensitive. Regex searches use PostgreSQL POSIX regular expression matching.
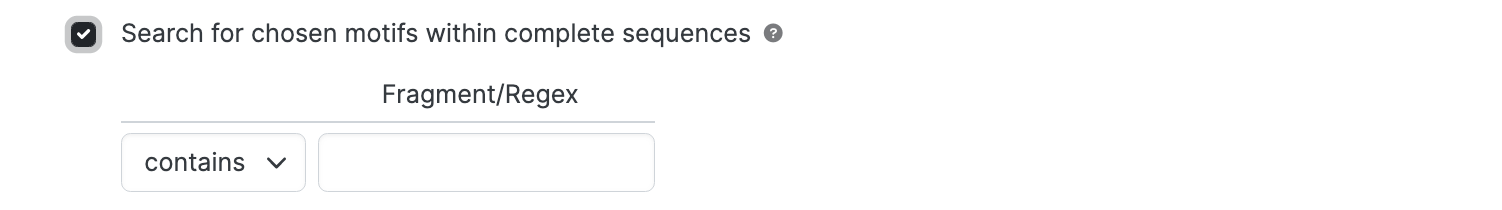
Search for chosen motifs within specific regions
 This is the same as for complete sequences but requires the selection of a numbered region.
This is the same as for complete sequences but requires the selection of a numbered region.
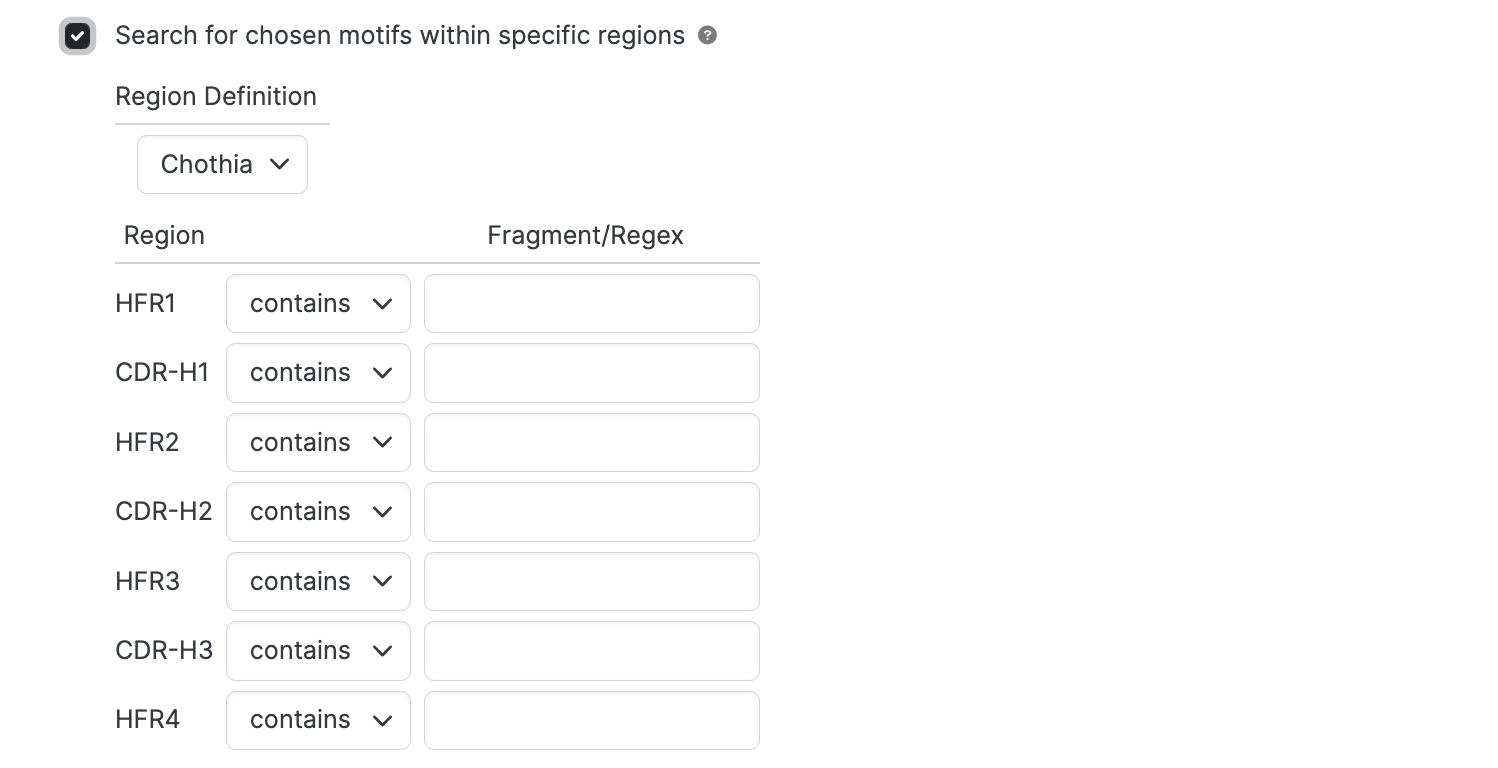
Specify minimum and maximum lengths for regions.
 This is useful when, for example, restricting a search to CDRs of particular loop lengths.
This is useful when, for example, restricting a search to CDRs of particular loop lengths.
For each region of interest:
- Use the Minimum dropdown to set the minimum length for the region.
- Use the Maximum dropdown to set the maximum length for the region.
Constrain amino acids at required positions
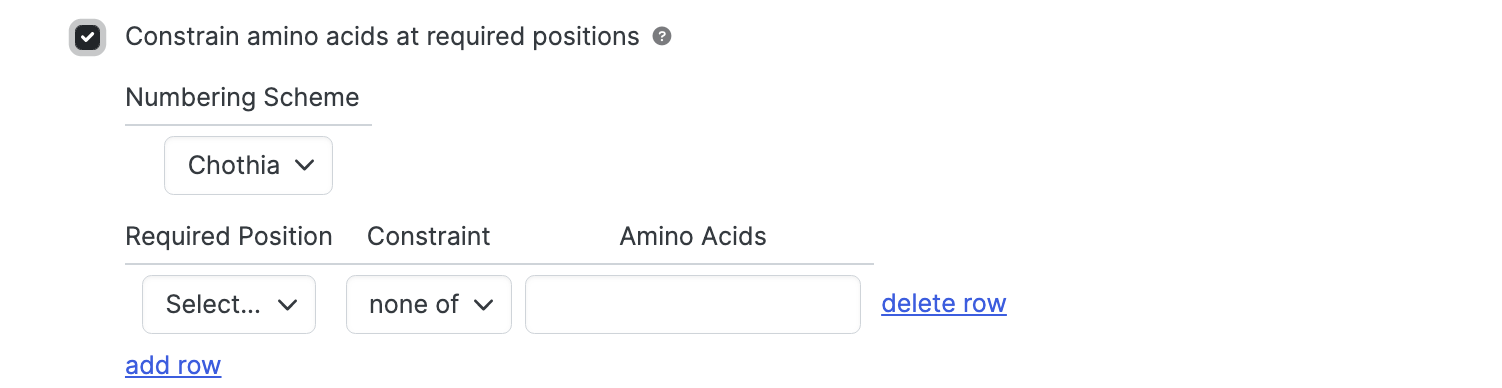
Use the Required Position dropdown to specify amino acids at particular positions to be present.
- Constraint select either one of or none of to control whether specified amino acids should be present or absent.
- Amino Acids specifies the amino acid types of interest using 1- letter code. List multiple amino acids with or without spaces or commas. Thus, SAV, S A V, or S,A,V will all specify serine, alanine, valine. The search is case insensitive.
- Add row for additional positions
- Delete row to remove.
If the Required Position dropdown is not selected in a given row, no constraint is applied and the row is effectively removed from consideration.
A chain will be included in the results only if all required positions are present and all the associated amino constraints are satisfied.
