-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Post Translational Modifications
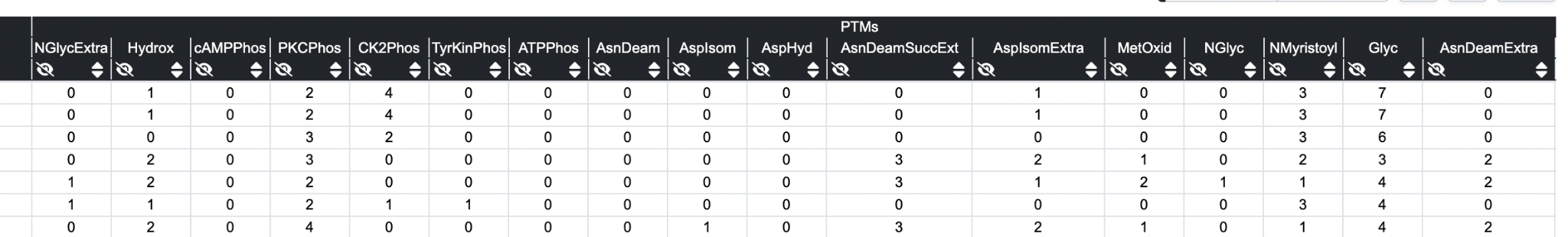
The following predicted post-translational modifications and characteristics are applied by regular expressions.
Note: that at the current time these are run on the whole sequence, not just the numbered region so a certain PTM might be recorded yet not displayed. Viewing an individual sequence on Key Annotation pages will restrict to the numbered region.
Lysine Hydroxylation (Hydrox)
Regex: '(?<=C.)([DN])(?=....[FY].C.C)'
This regular expression defines twelve residues in which the first must be C, the third must be either D or N, the eighth must be either F or Y, and the tenth and twelfth must both be C.
PKC phosphorylation site (PKCPhos)
Regex: '([ST])(?=.[RK])'
This regular expression defines three residues in which the first must be either S or T and the third must be either R or K.
Asparagine deamidation site (AsnDeam)
Regex: '(N)(?=G)'
This regular expression defines two residues which must match the pattern N followed by G.
Methionine oxidation (MetOxid)
Regex: '(M)'
This regular expression indicates the occurrences of the residue M.
N-linked glycosylation site (NGlyc)
Regex: '(N)(?=[^P][ST][^P])'
N-linked glycosylation site extended (NGlycExtra)
Regex: '(N)(?=.[STC])'
This regular expression defines three residues in which the first must be N and the third must be either S, T or C.
cAMP phosphorylation site (cAMPPhos)
Regex: '(?<=[RK][RK].)([ST])'
This regular expression defines four residues in which the first must be either R or K, the second must be either R or K, and the fourth must be either S or T.
CK2 phosphorylation site (CK2_Phos)
Regex: '([ST])(?=..[DE])'
This regular expression defines four residues in which the first must be either S or T and the fourth must be either D or E.
Tyrosine Kinase Phosphorylation site (TyrKinPhos)
Regex: '[RK].{2,3}[DE].{2,3}(Y)'
This regular expression defines between seven and nine residues in which the first must be either R or K; this followed by two or three of any residue. The next residue must be either D or E, followed by two or three of any residue. The final residue must be a Y.
ATPASE E1 E2 phosphorylation site (ATPPhos)
Regex: '(D)(?=KTGT[LIVM][TI])'
This regular expression defines seven residues where the first five must be DKTGT, the sixth must be either L, I, V or M, and the seventh must be either T or I.
Aspartate isomerization site (AspIsom)
Regex: '(D)(?=G)'
This regular expression defines two residues which must match the pattern D followed by G.
Aspartate hydrolysis site (AspHyd)
Regex: '(D)(?=P)'
This regular expression defines two residues which must match the pattern D followed by P.
Asparagine deamidation succimide extended (ASN_DEAMIDATION_EXTRA)
Regex: '(N)(?=[SATND])'
This regular expression defines two residues in which the first must be N and the second must be either S, A, T, N or D.
Aspartate isomerization site extended (AspIsomExtra)
Regex: '(D)(?=[PSNHD])'
This regular expression defines two residues in which the first must be D and the second must be either P, S, N, H or D.
N-Myristoylation (N myristoyl)
Regex: '(G)(?=[^EDRKHPFYW]..[STAGCN][^P])'
Glycation (Glyc)
(K)
Asparagine Deamidation Extended (AsnDeamExtra)
Regex: '(N)(?=S)'
Glutamine Deamidation Extended (GluDeamExtra)
Regex: '(Q)(?=G)'
