-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Structure
When Coordinates are available use this Tab to interrogate 3D structure
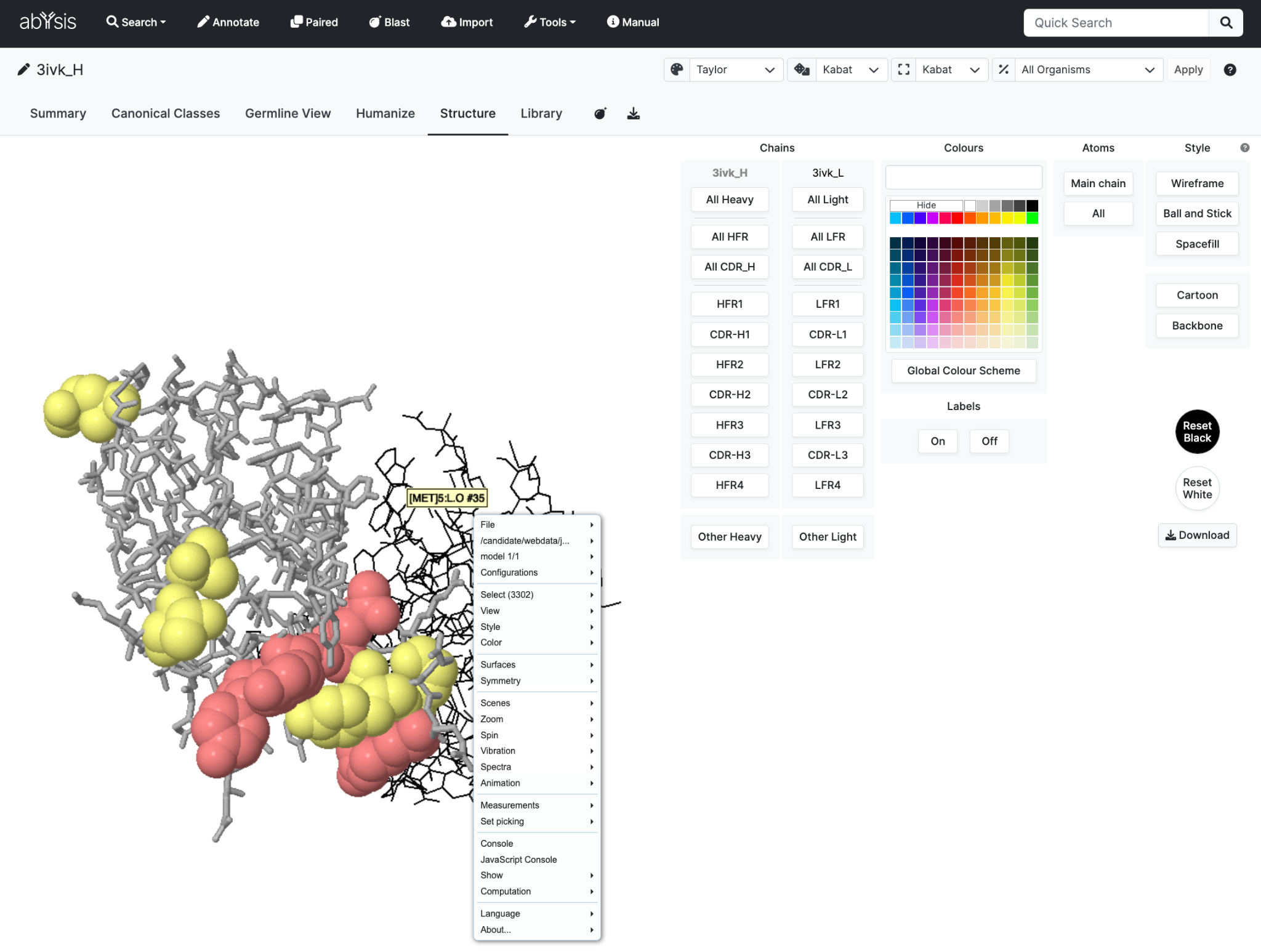
The interface combines dedicated options to integrate with abYsis especially with the Humanise Tab as well as allowing existing JSMol options to be used from standard menus.
Basic Controls
To rotate the structure, simply click on the structure with the (usually left) mouse button and move it around.
To rotate the structure in the z-axis (i.e. the axis coming out of the screen), press and hold the shift button, click the left mouse button and move the mouse left and right.
To zoom the structure either use the scroll on the mouse, or press and hold the shift button, click the left mouse button and move the mouse up and down.
Display controls
All chains in a structure file are loaded but they might not all be displayed.
For each chain, there is a table of display controls.
Select numbered CDR regions of interest and colour based on user selection or the current Global Colour Scheme. This can be very powerful when colouring by frequency data to highly unusual residues.
Control how much of each amino acid residue is displayed by selecting Atoms (Main Chain or All) and Style (Wireframe, Ball and Stick or Spacefill).
Two macro settings are also available; Cartoon and Backbone which combine Atoms with Style.
Reset will allow you to choose either a white or black background.
Download is for a PDB coordinate file.
