-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Overview
Let’s quickly introduce the 3 key ways to analyse your set of sequences.
Annotate
For quickly analysing up to about 100 sequences, use Key Annotate (& make sure only amino-acid or DNA are done in a single search, not a combination).
abYsis will quickly:
- Number your sequences
- Assign Framework and CDRs
- Predict Post Translational Modifications
- Predict Canonical CDR conformations
Here we’ll demonstrate with 5 DNA sequences (DNA is more challenging as it needs to find the reading frame with the antibody sequence).
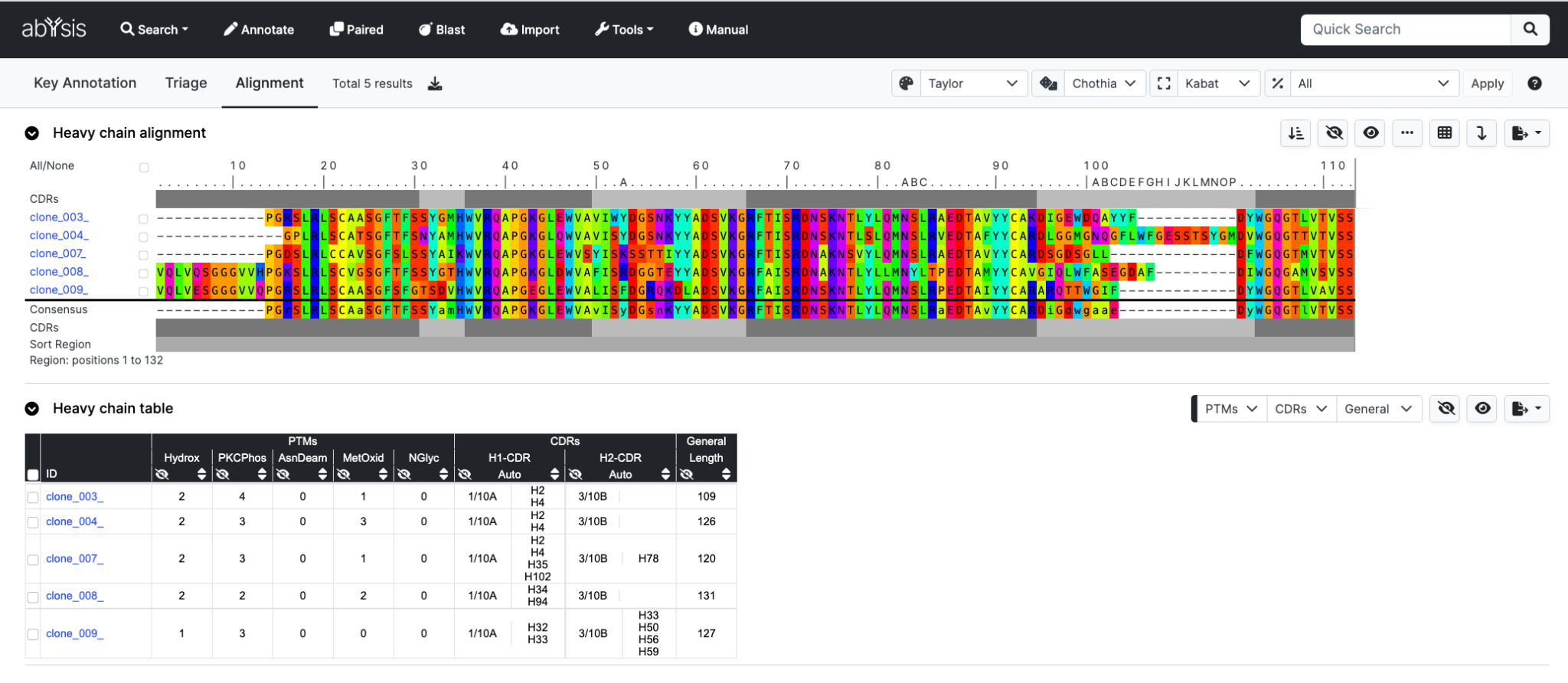
And here’s the output
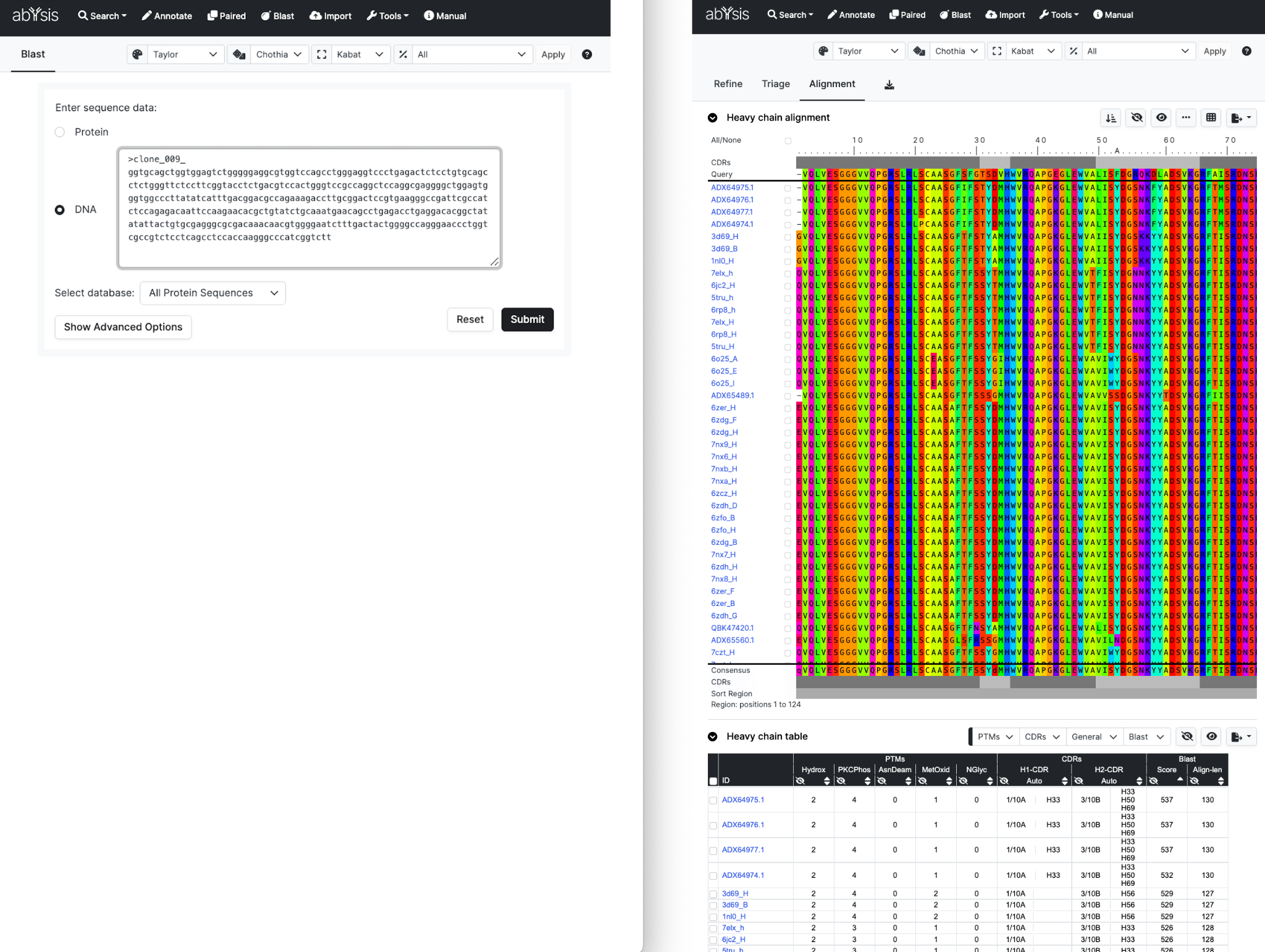
Blast
You can perform a BLAST search for a single query (protein or DNA) against proteins in abYsis. Here is a simple example.
There’s various opportunities to add/remove columns in the data Table and as well as to restrict the databases scanned by Blast.
Import
If you have too many sequences to analyse interactively, or want to use the same sequences every day without having to recalculate, use the Import option.
Your sequences will be processed and retained in your version of abYsis. Import is a great choice up to a maximum of 10,000 sequences in a single upload.
Upload your file, ensuring that it is in fasta format and that the header line for each sequence has the necessary 6 fields, even if some are left blank. See detailed section on Import for more information.