-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Global Selectors

Throughout the system, the default choice of Sequence Colour, Numbering Scheme, CDR Definition and Organism is controlled using the Global Selector.
To change the defaults for your session, select from each dropdown box and click 'Go'. The new desired combination will be maintained for your session.
In the following image, our alignment is now displayed by position dependent amino-acid frequency data calculated over all organisms. 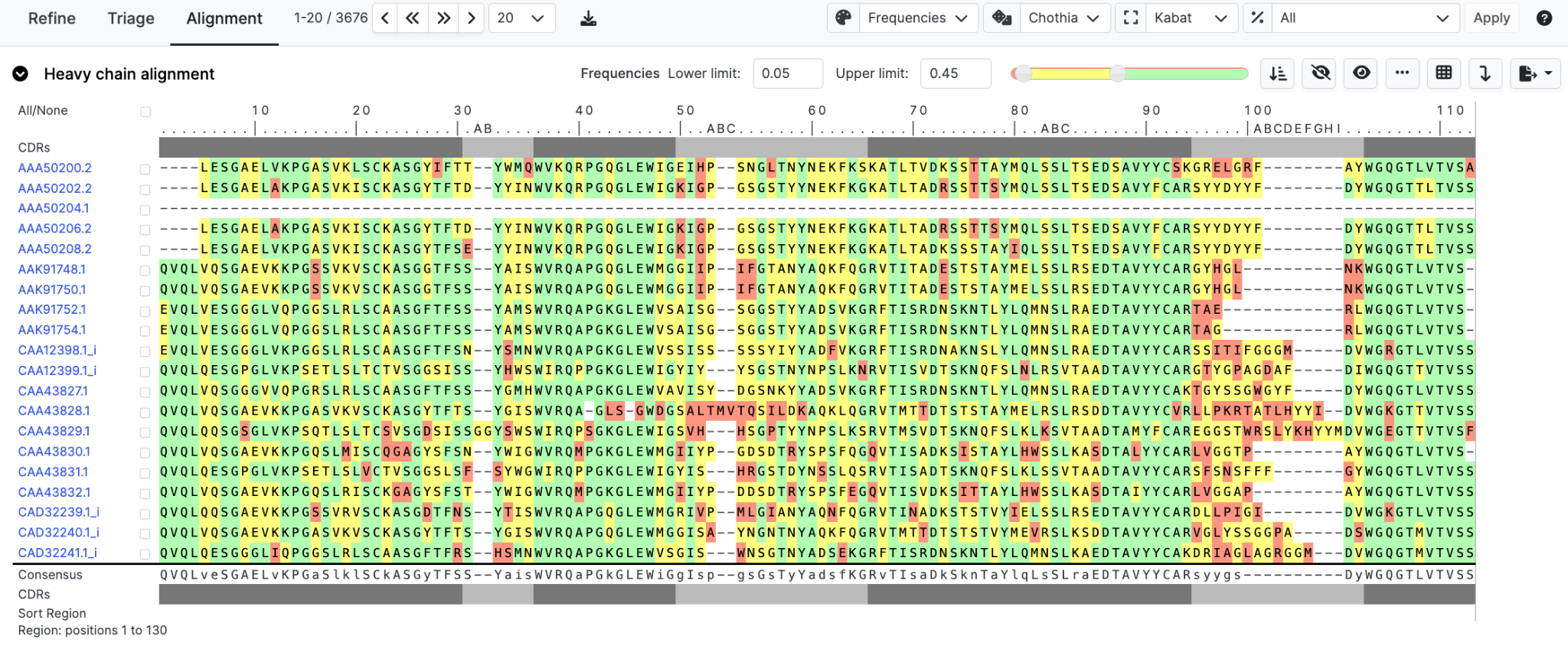
To restrict frequencies to a desired organism use the dropdown menu
Refine Triage Alignment
For pages with multiple results you will typically see three options including the default Alignment output. The other two are Triage, to filter down large sets to more management numbers of sequences and Refine where you can adjust your query.

Refine is essentially the search page prepopulated with the existing query whereas Triage output looks like the following figure with interactive selection of CDR and Framework regions based on length as well as Post Translational Modifications - see detailed sections in the manual to learn how these work.