-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Annotate
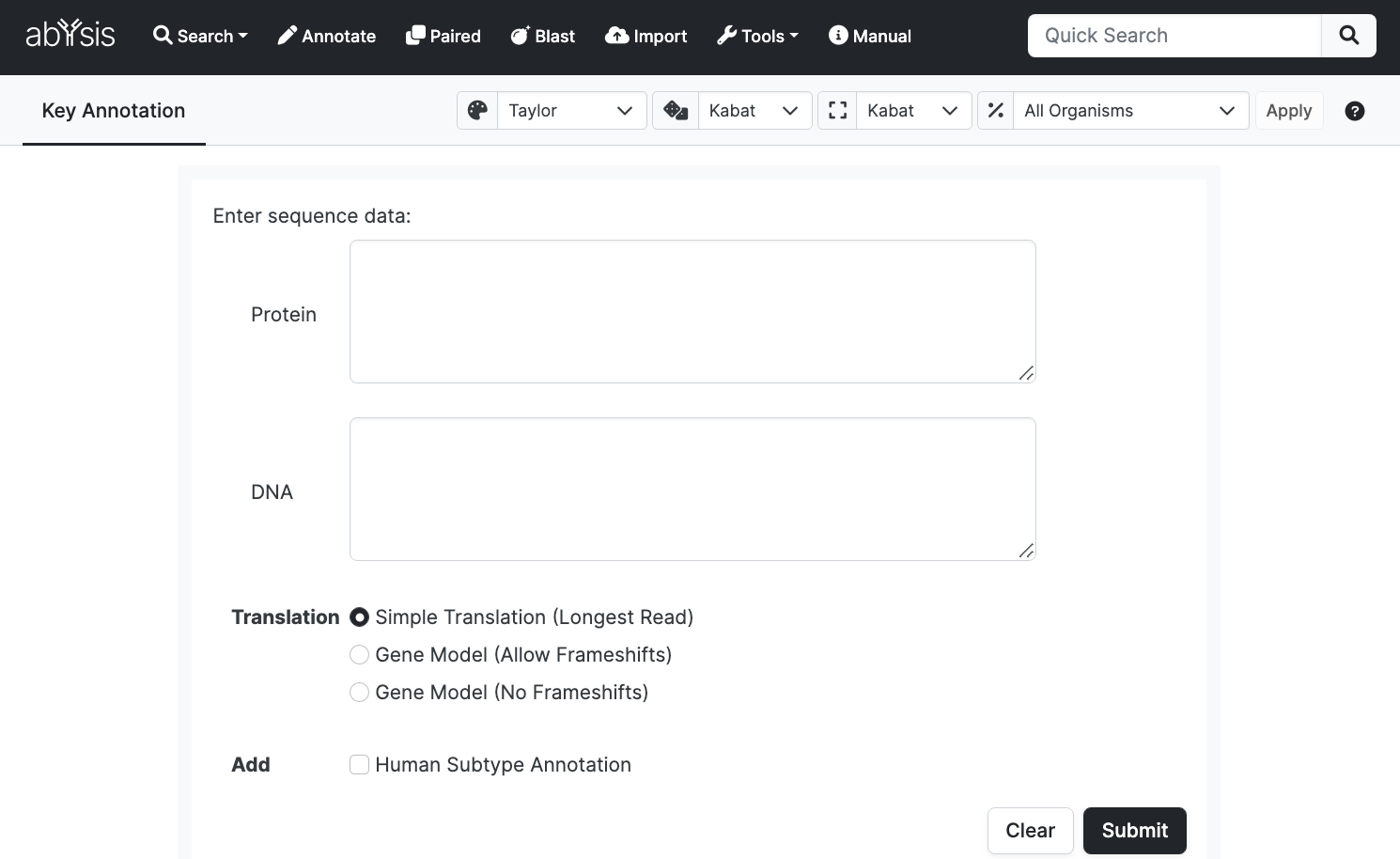
Single Entry
For a single antibody, a DNA sequence or a Protein sequence may be entered.
If only a Protein sequence is entered the system will simply use the sequence entered.
If only a DNA sequence is submitted, abYsis will attempt to translate the DNA according to the option selected.
One of three Translation methods can be selected:
- Simple Translation (Longest Read) (default): Examines all 6 reading frames and selects the longest uninterrupted amino acid read. For the majority of research this is the most useful.
- Gene Model (Allow Frameshifts): Compares user DNA sequence to Germline DNA for V D J regions. Permits frameshifts in both user and target DNA in order to generate a long amino acid read. This is useful if possibility that user input has sequencing errors but can lead to erroneous Germline assignments by permitting further indels.
- Gene Model (No Frameshifts): Allows no frameshifts in user or Germline DNA. Translated amino acid sequence will most commonly be V region only.
Exceptionally, a pair of sequences, one DNA sequence and one Protein sequence can be entered. In this case abYsis will just align the DNA to the declared protein translation rather than generate a protein translation from the DNA. Note that the header must be identical if used.
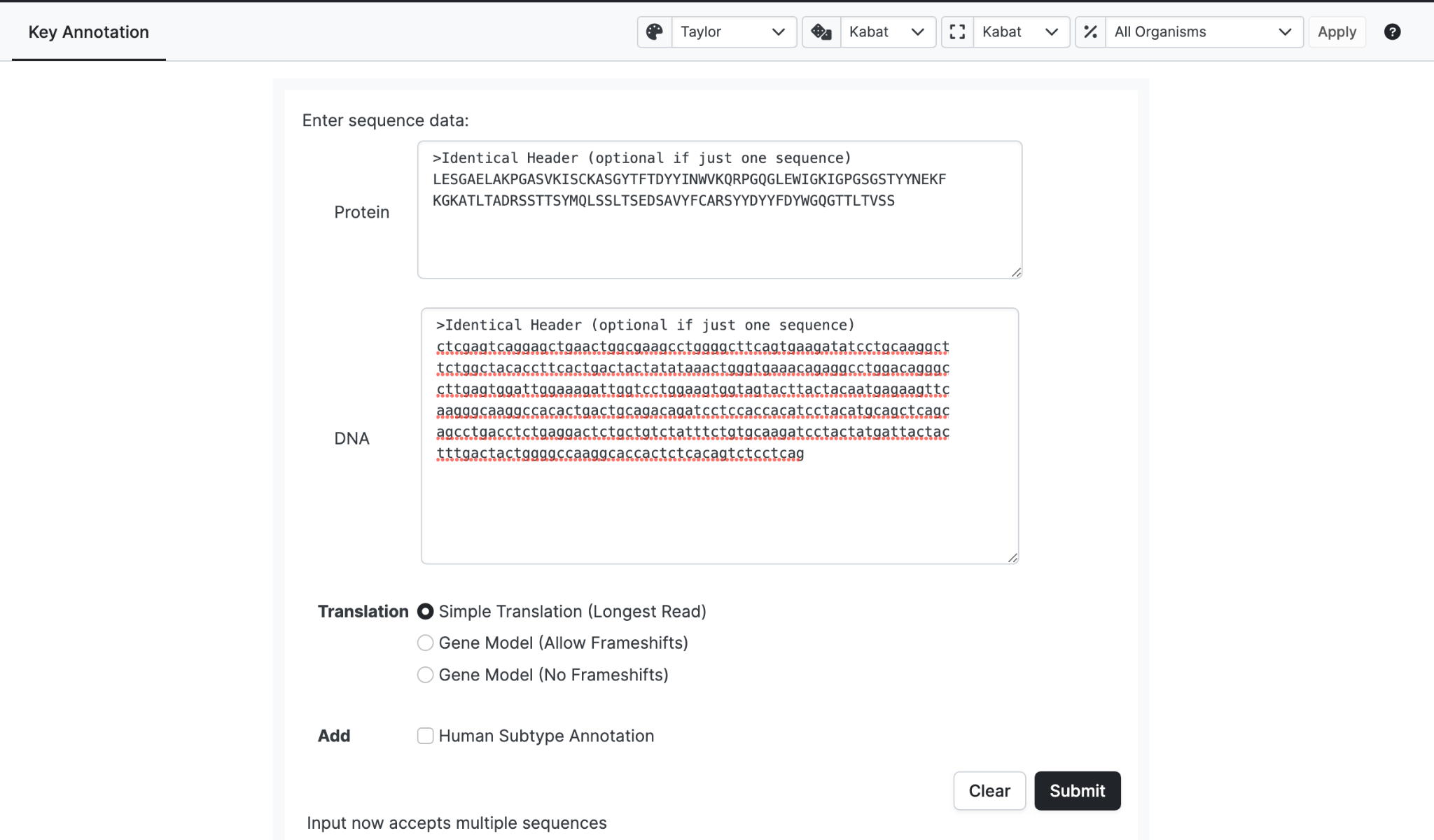
If your organism is Human you may select the Human Subtype Annotation checkbox. 
(Note that if you select and use for non-human sequences, results will still be provided but are not meaningful).
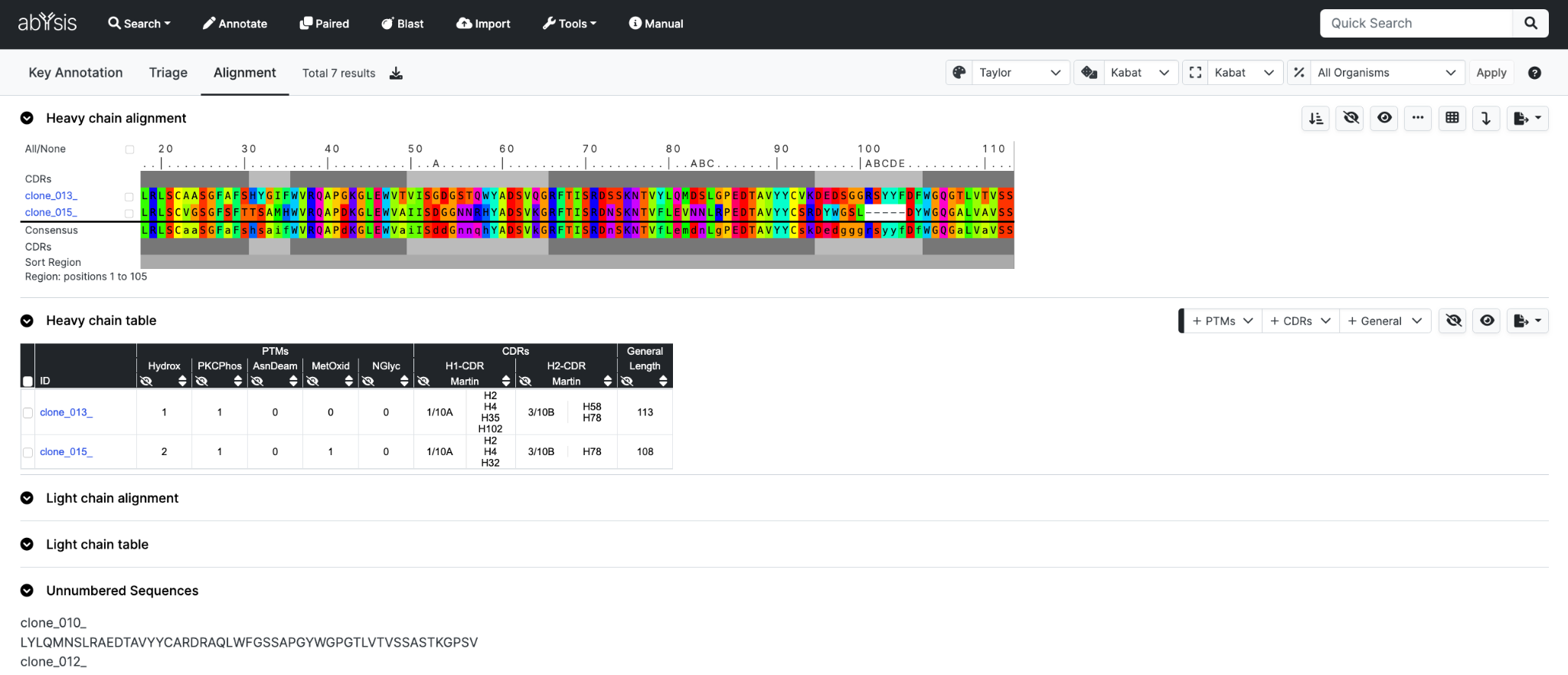
For a single entry you will be taken straight to the following pages.
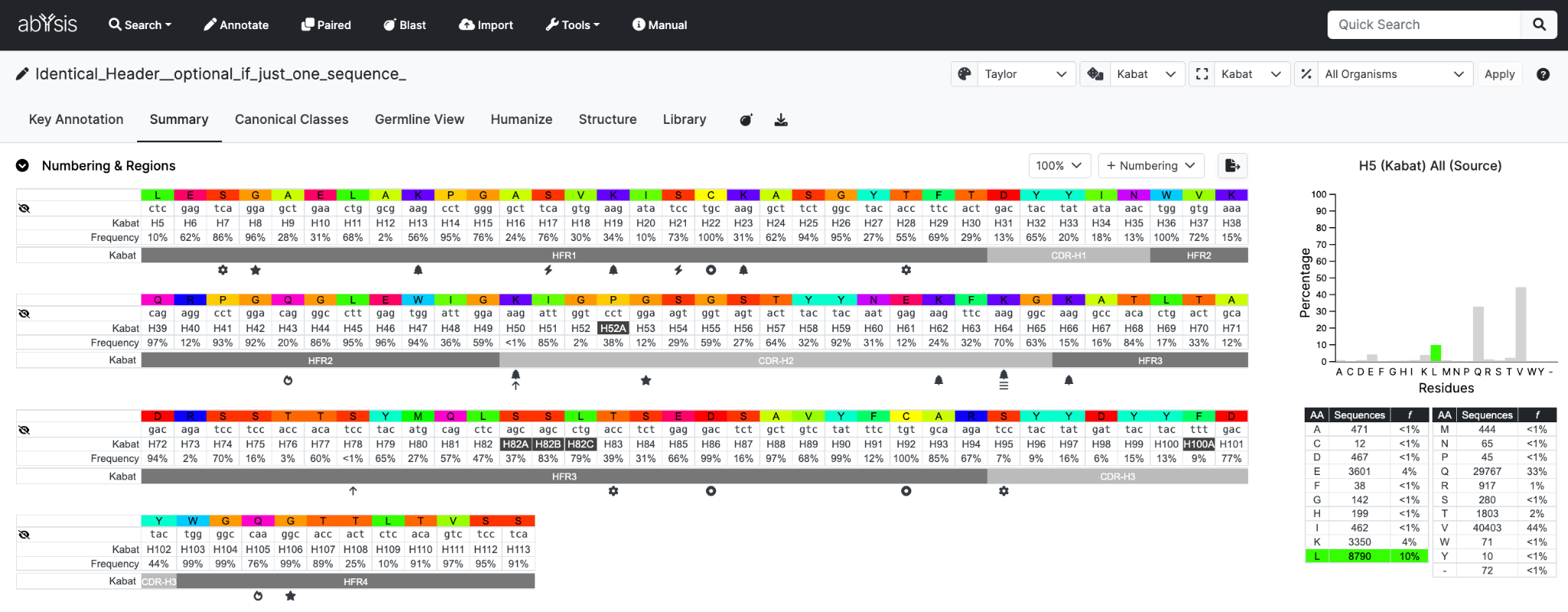
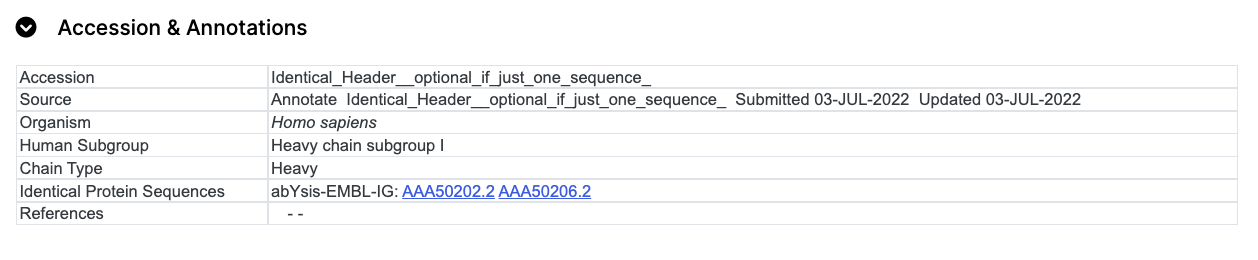
The Human Subgroup option, if selected, will be output in the Accession & Annotations table;
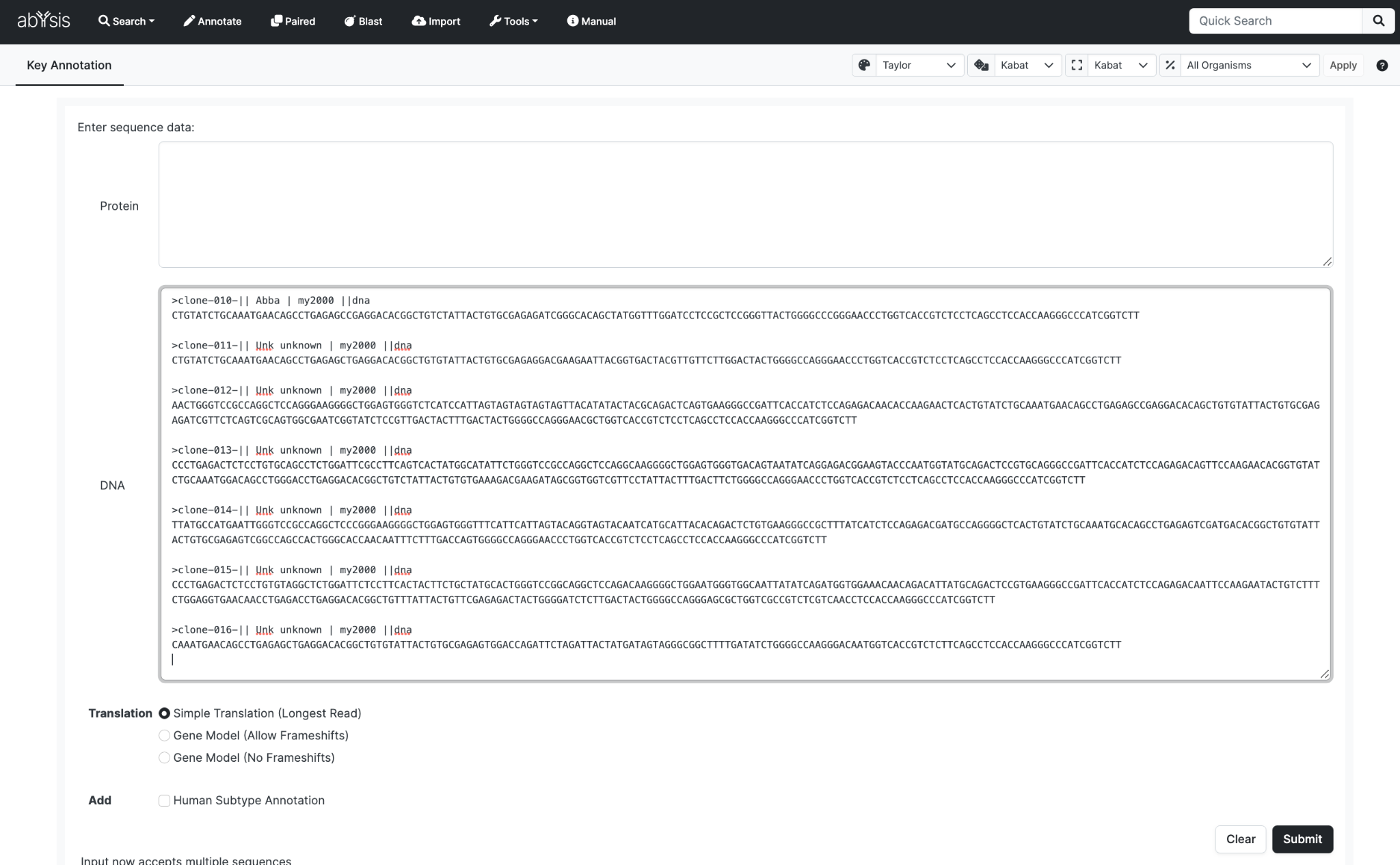
Multiple Entries
These can be entered using a FASTA format where each antibody must have a unique FASTA identifier.
We recommend that for a single run you only use either Protein sequences or DNA sequences, not both.
If both DNA and Protein sequences are submitted for a particular entry, the FASTA identifiers for the equivalent DNA and Protein sequence must match and the order of the entries must match.
For multiple sequences the initial output with be the numbered alignment. Sequences that are not successfully numbered most likely due to being too short in length, will not be displayed in the alignment.