-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Paired Analysis
New for v4.0
Whilst the multiple alignment page of abYsis shows paired Heavy and Light chains, other pages have always treated each Heavy or Light chain individually.
Paired Analysis now allows you to analyse chains using a tailored version of the Humanize facility, all on a single page. Where available, 3D structure can also be analysed.
Either extract data from the database using Accession code (specify Data Source if two files with same code) or load 2 sequences to Annotate together.
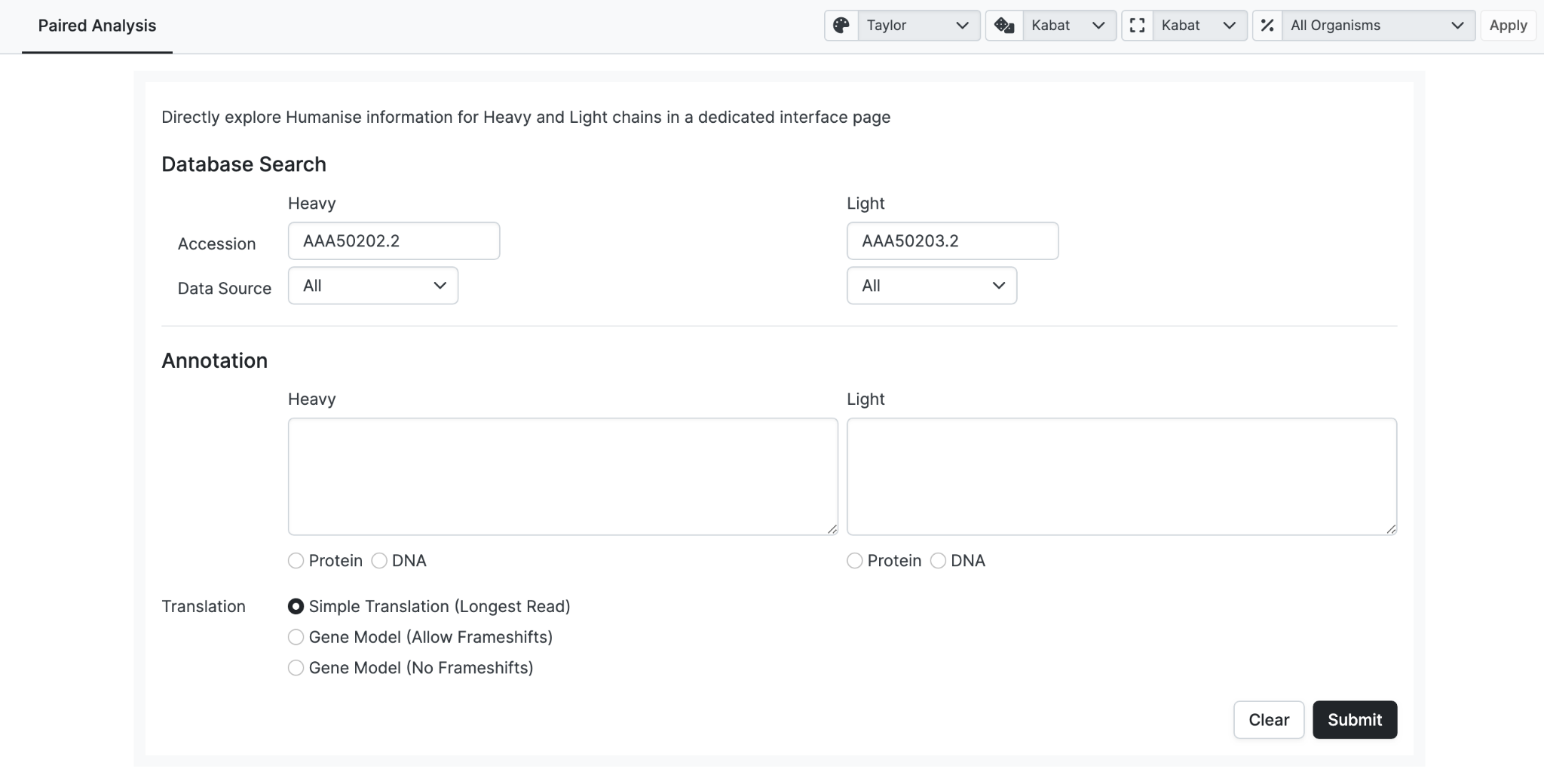
Here we enter Heavy and Light chains that are already in the database
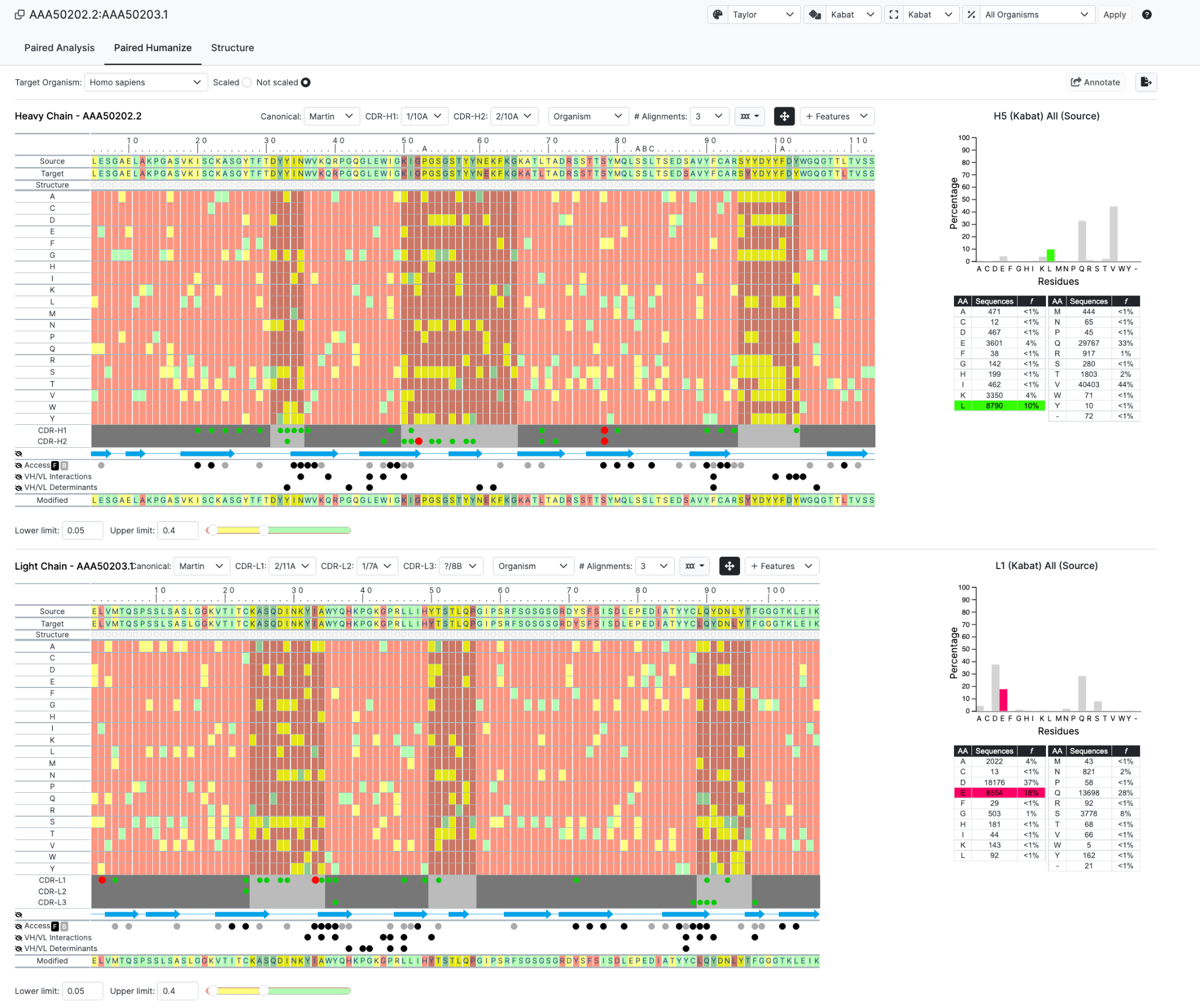
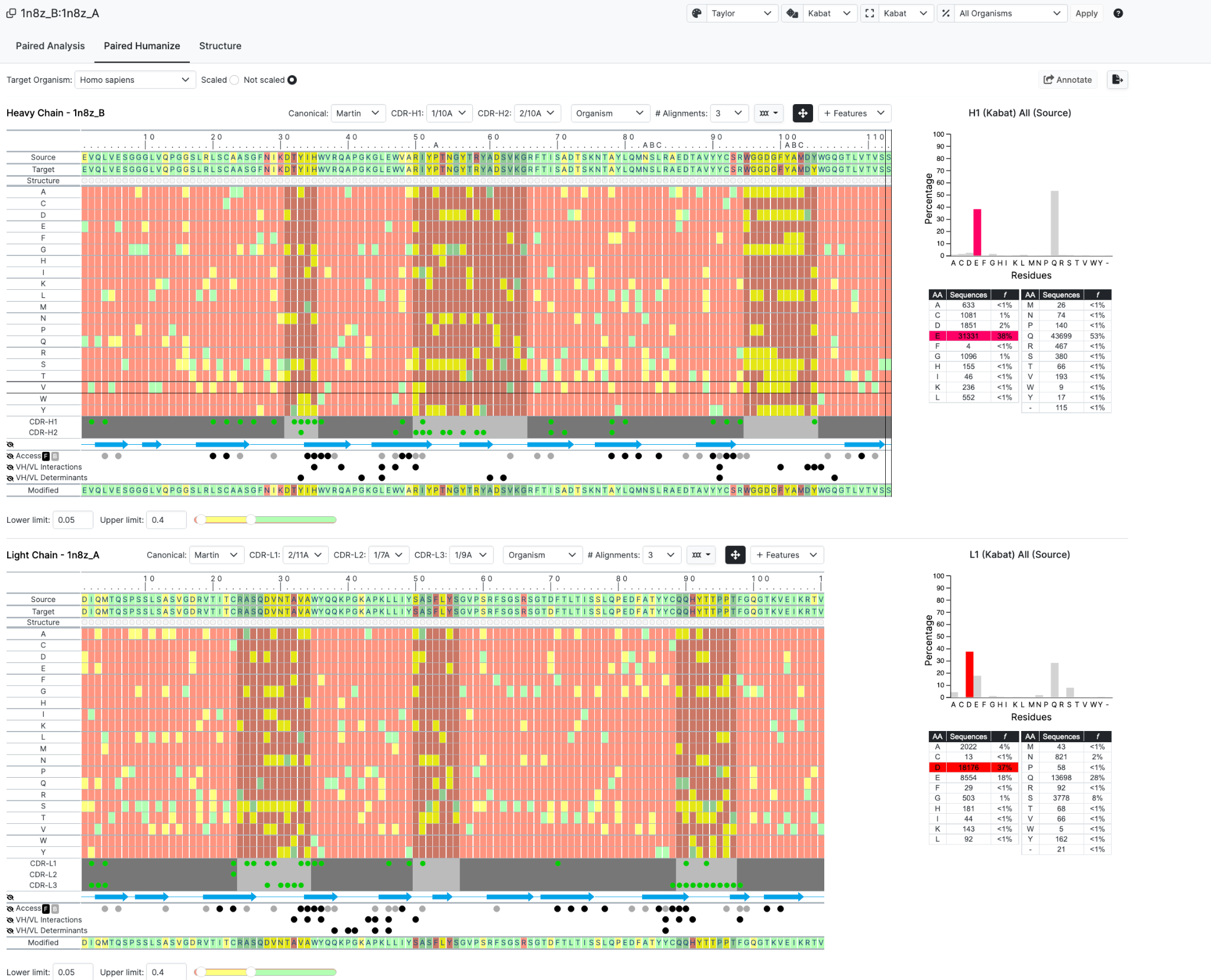
In this new Paired interface you are now taken straight to the Humanize layout but for both chains. All germline and and canonical information is provided but without the individual tabs that you would get with accessing a chain at a time.
Going to the Structure tab will show this message;
We have plans to introduce a Homology Modeling option to allow people to build models of their sequences of interest. However for the time being, if you have access to other homology modeling packages you could load combined sequence/3D coordinates in PDB format through the Import facility.
However for this example we will take an experimental coordinates for PDB entry to show functionality.
First we found the codes of interest through the basic search by looking for Name; herceptin and Data Source; PDB. From that search we identified file 1n8z as being of interest.
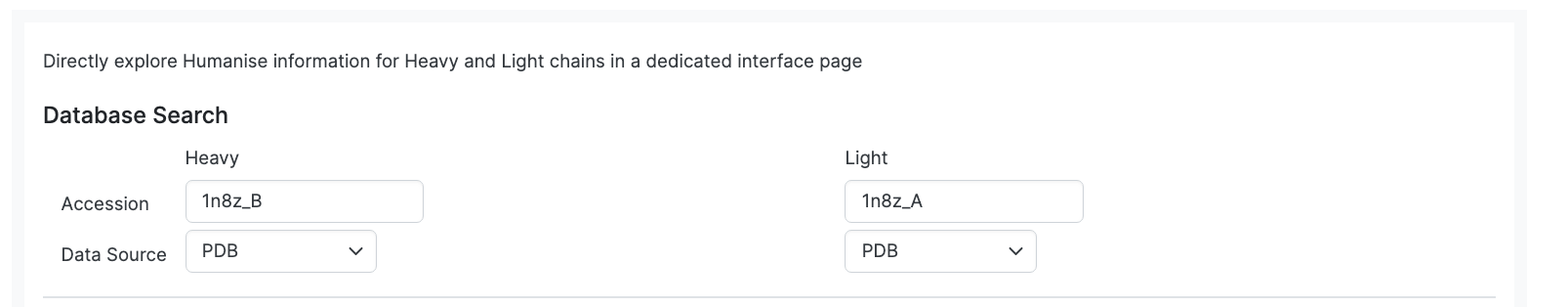
Once submitted we see the following page of the Paired Humanize
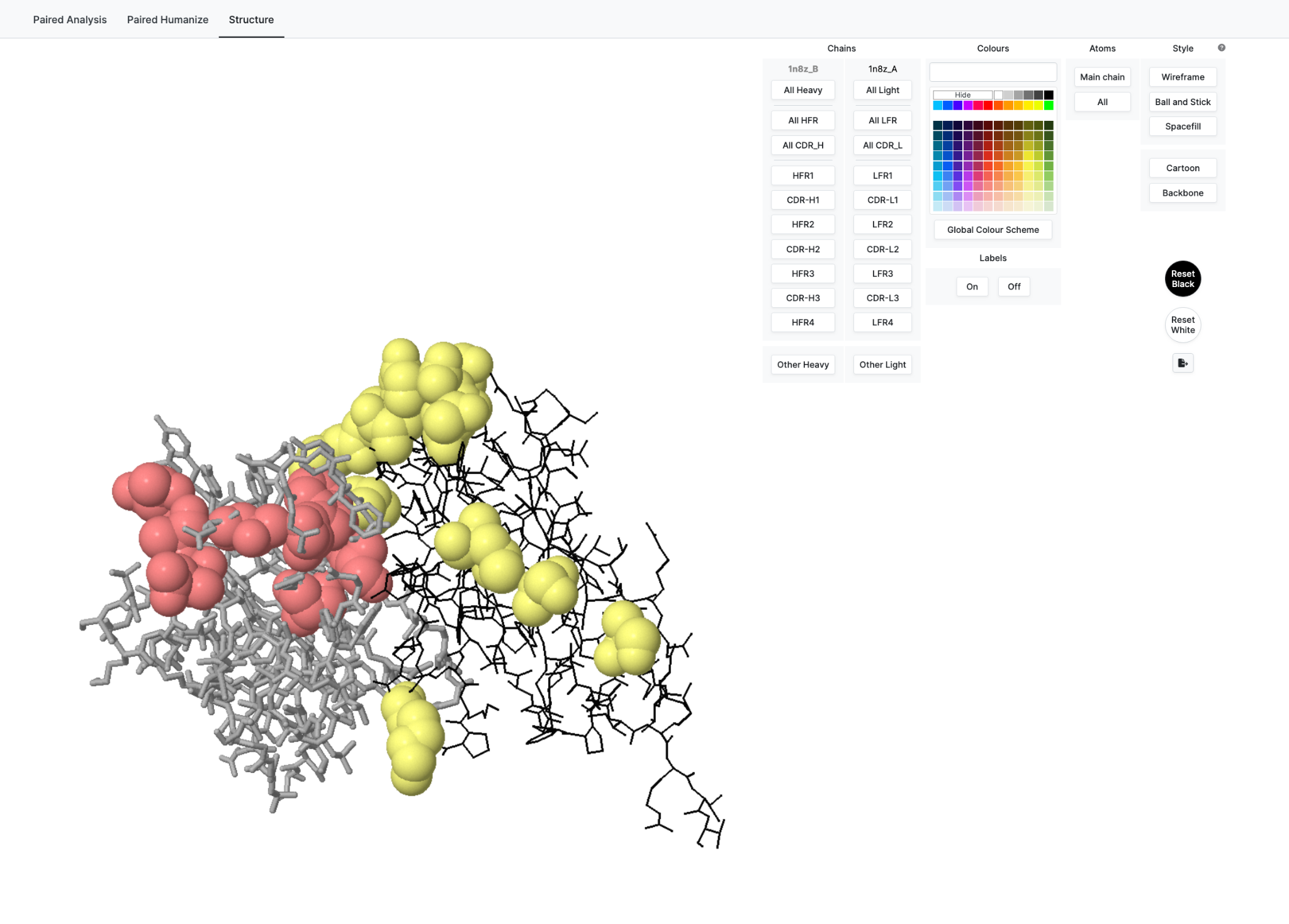
And we see 3D structure on the Structure Tab
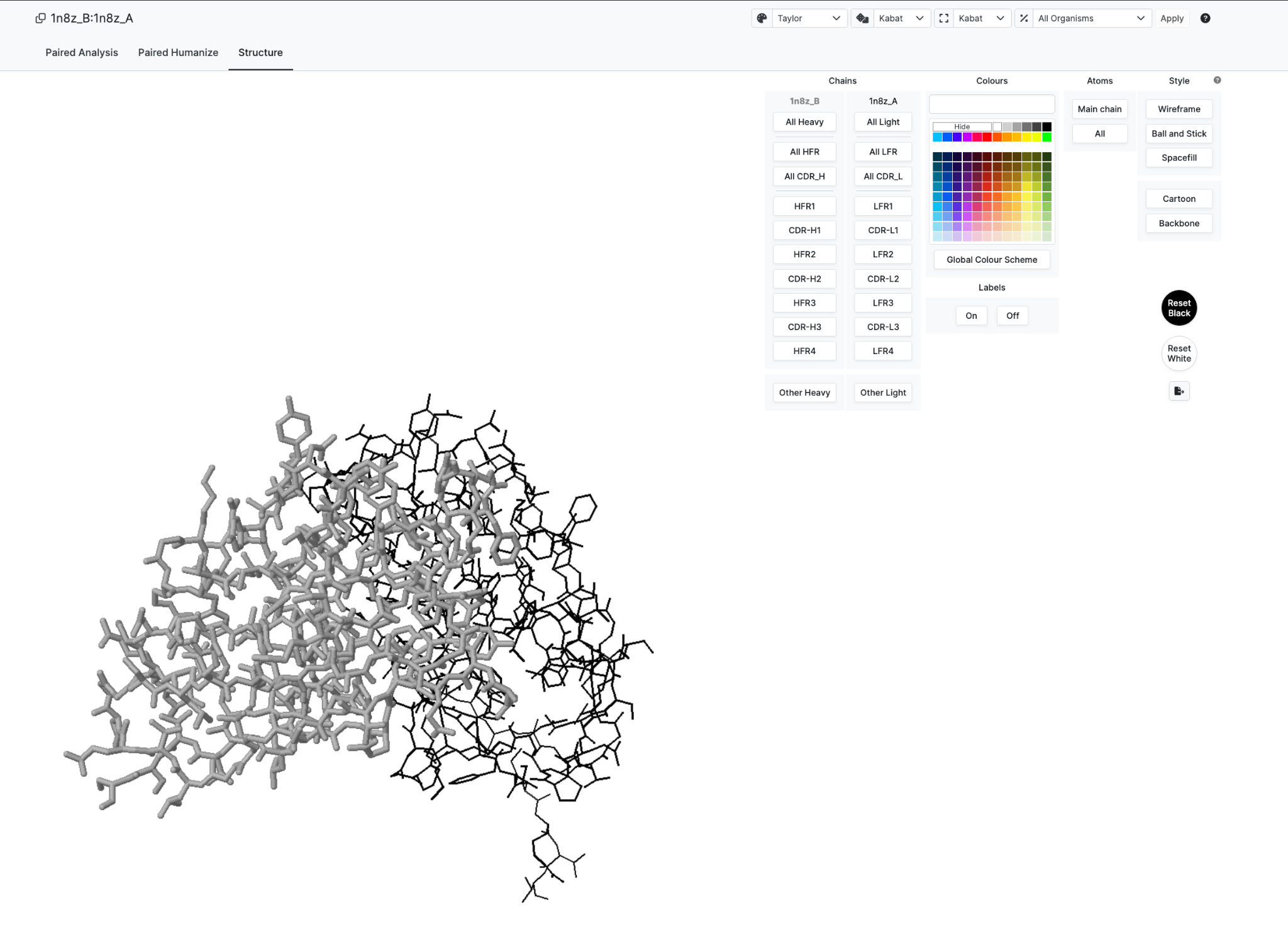
Now to introduce the functionality we have selected certain residues to highlight in Red for Heavy Chain and Yellow for light Chain (see Structure Row just below Sequence)
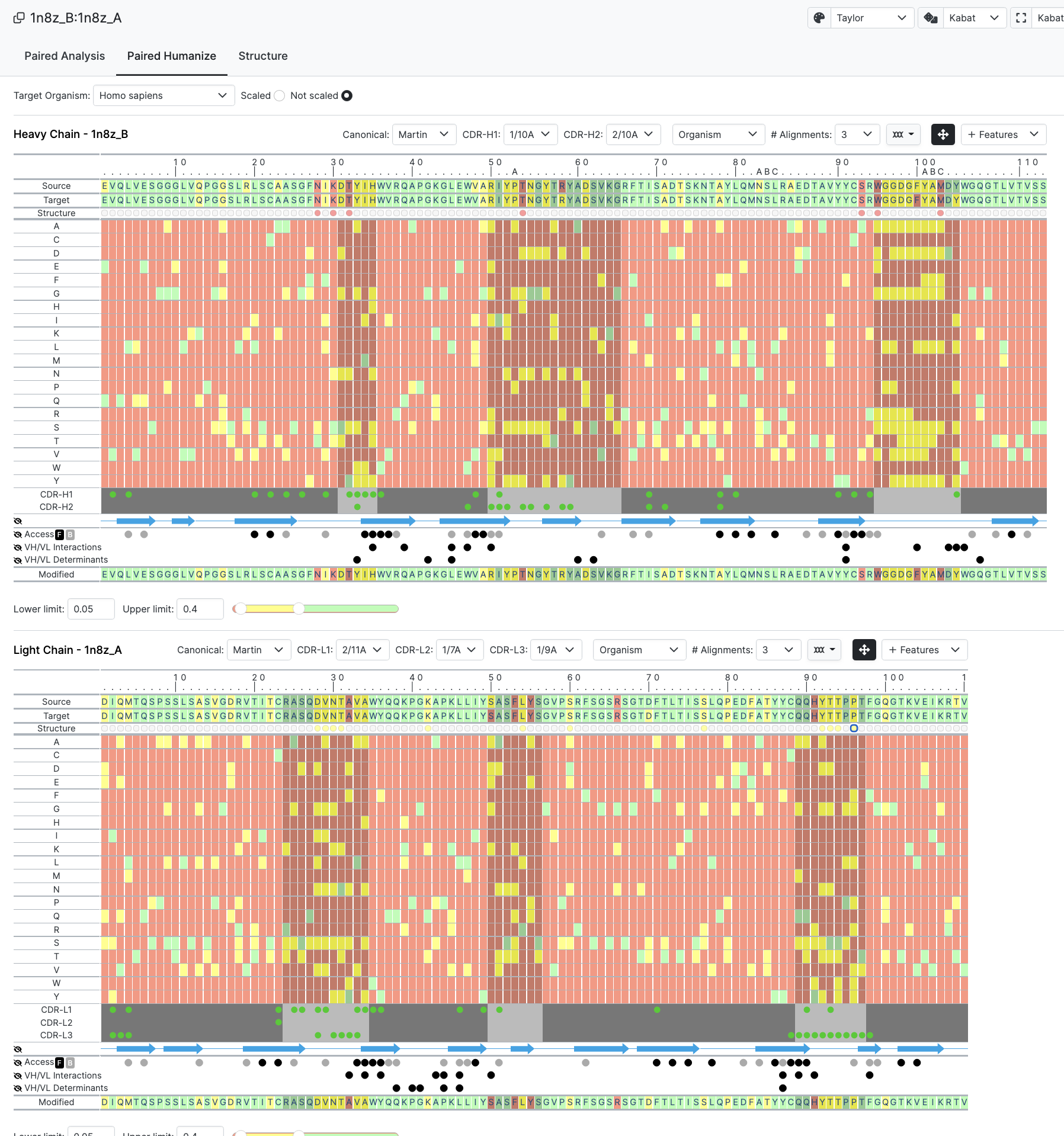
Selecting these immediately communicates the selection to the 3D structure tab allowing seamless analysis.