-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Amino Acids & Regions
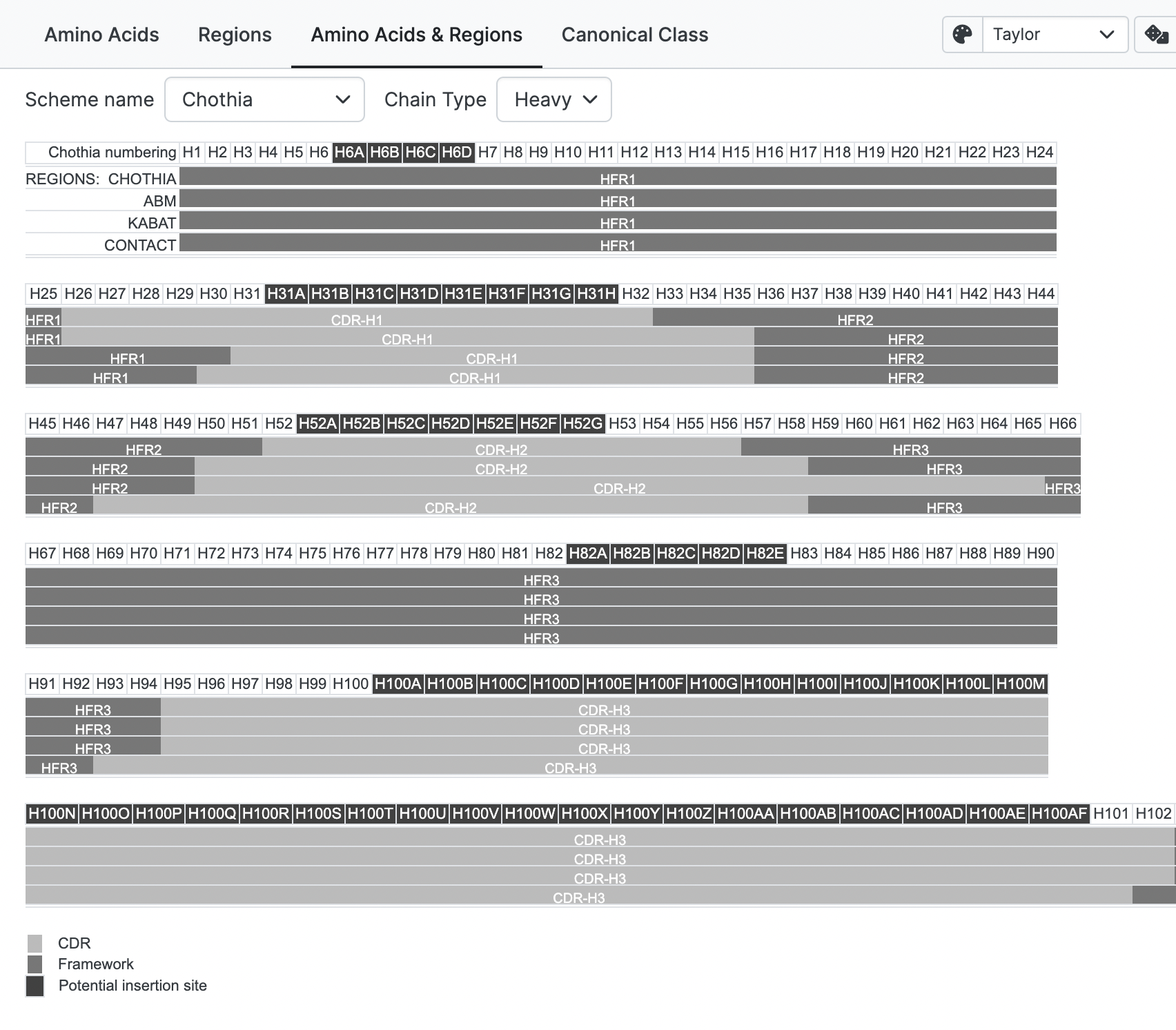
Numbering
Five numbering schemes are available:
- Kabat: Most widely adopted standard but insertions occur in CDR-L1 and CDR-H1 do not match the real structural insertion positions.
- Chothia: similar to the Kabat but places CDR-L1 and CDR-H1 insertions at the structurally correct positions.
- Martin: enhanced version of the Chothia scheme that additionally places Framework insertions and deletions at structurally correct positions.
- IMGT: Implementation of an IMGT numbering scheme. Pre-defines theoretically longest possible loops
- Aho: Structurally defined numbering. Similar to IMGT but ensures structurally accurate insertion and deletion positions.
CDR/Framework Scheme
Five CDR definitions are available:
- Kabat, based on analysis of sequence variability.
- Chothia, based on the location of 3D structural loop regions. (Note that Chothia definitions varied between different papers – this is the one judged to be most relevant.
- AbM Compromise between the Kabat and Chothia. Whitelegg N & Rees AR, Protein Eng. 13(2000):819-824 Methods Mol Biol. 248(2004)51-91).
- Contact: Based on an analysis of which residues contact antigen in crystal structures. (MacCallum RM, Martin ACR & Thornton JM, J. Mol. Biol. 262(1996)732-745).
- IMGT: CDR and Framework definitions as defined by IMGT
Note
Chothia region definition for CDR-H1 loop, and Contact definition for CDR-L1 loop, have a complex specification when using the Kabat numbering scheme. However, for a given sequence, the CDR regions are ultimately independent of the numbering scheme. Internally, abYsis prioritises the Chothia numbering scheme to delineate regions.
