-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Advanced Table Display
When viewing results it is possible to add further annotation. These are divided into Sequence Detail and Structure Detail.
Note: Do not get these confused with the similarly named Search functions.
Sequence Detail
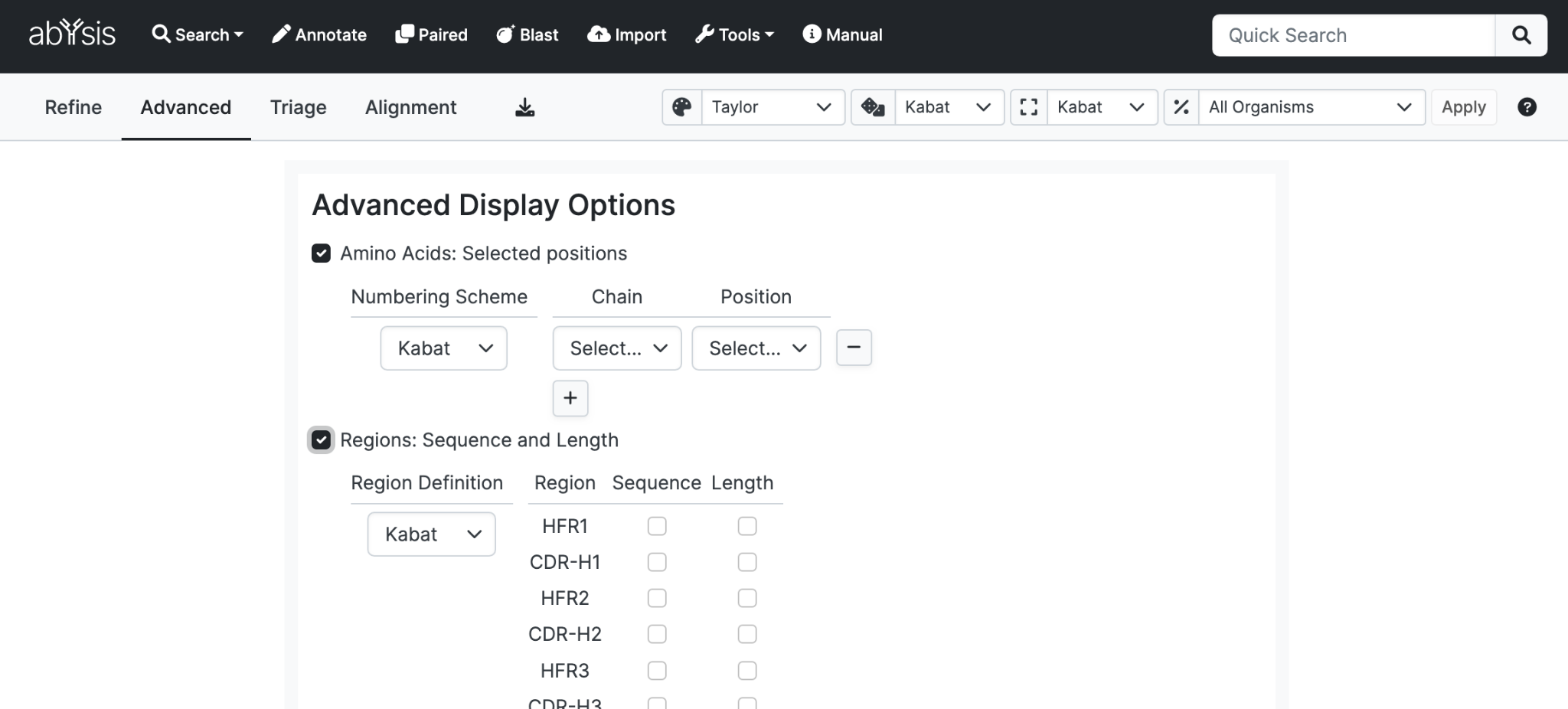
Display annotation corresponding to the Sequence search criteria in the query form.
- Amino Acids: Selected positions. Specify the Chain, Numbering Scheme and the residue Position using that scheme to display a column showing the residue present at the selected position. For example, show the residue position at Kabat position H23 for heavy chains.
- Regions: Sequence and Length. Specify the Region Definition (Chothia, Kabat, AbM or Contact) and check the boxes of what you want to display - Sequence and/or Length for each of the regions. For example, you could add a column showing the sequence for the Kabat CDR-L3 region in each hit.
Structure Detail
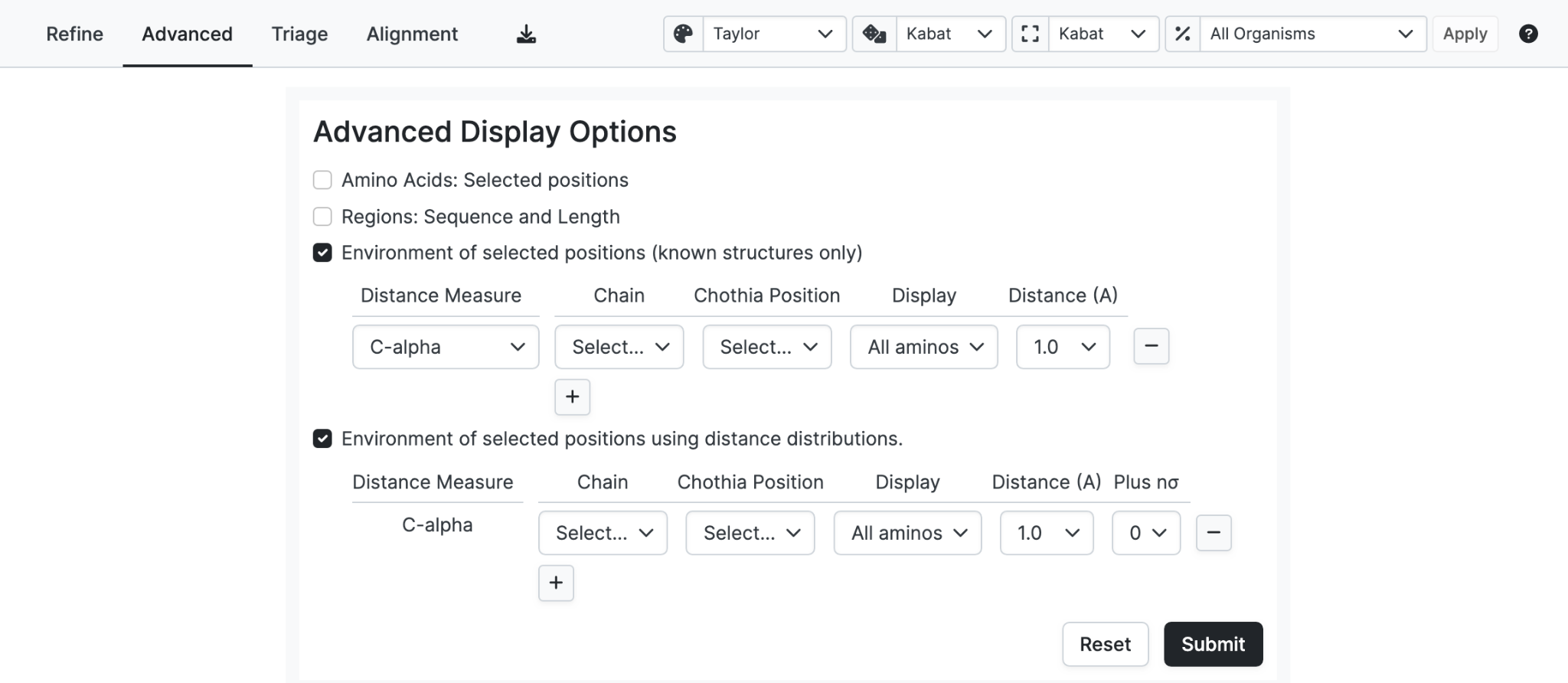
Display annotation corresponding to the Structure search criteria in the query form.
- Environment of selected positions (known structures only): For the selected chain, shows specified residues within a given distance in known structures. For example, show all tryptophan (W) and tyrosine (Y) that have a closest atom within 6.0Å of L32 in light chains.
- Environment of selected positions using distance distributions: For the selected chain, shows specified residues within a given distance in known structures and in sequences using predicted distances. For example, show all light chain residues that have a C-alpha to C-alpha distance of ≤6.0Å from L32.